+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-24258 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of the in situ yeast NPC | ||||||||||||||||||||||||
 マップデータ マップデータ | full in situ NPC map recombined from 90 degree wedge focused alignment | ||||||||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||||||||
 キーワード キーワード | NPC / nucleocytoplasmic transport / TRANSLOCASE | ||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報response to spindle checkpoint signaling / nuclear pore linkers / peroxisomal importomer complex / regulation of protein desumoylation / mRNA export from nucleus in response to heat stress / nuclear pore inner ring / Seh1-associated complex / positive regulation of ER to Golgi vesicle-mediated transport / protein exit from endoplasmic reticulum / COPII-coated vesicle budding ...response to spindle checkpoint signaling / nuclear pore linkers / peroxisomal importomer complex / regulation of protein desumoylation / mRNA export from nucleus in response to heat stress / nuclear pore inner ring / Seh1-associated complex / positive regulation of ER to Golgi vesicle-mediated transport / protein exit from endoplasmic reticulum / COPII-coated vesicle budding / protein localization to nuclear inner membrane / chromosome, subtelomeric region / COPII-mediated vesicle transport / transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery / nuclear pore localization / adenyl-nucleotide exchange factor activity / nuclear pore central transport channel / regulation of nucleocytoplasmic transport / establishment of mitotic spindle localization / nuclear migration along microtubule / regulation of TORC1 signaling / nuclear pore organization / nuclear pore outer ring / nuclear pore complex assembly / telomere tethering at nuclear periphery / COPII vesicle coat / post-transcriptional tethering of RNA polymerase II gene DNA at nuclear periphery / positive regulation of protein exit from endoplasmic reticulum / nuclear pore cytoplasmic filaments / Transport of Mature mRNA derived from an Intron-Containing Transcript / Regulation of HSF1-mediated heat shock response / nuclear pore nuclear basket / tRNA export from nucleus / SUMOylation of SUMOylation proteins / cytoplasmic dynein complex / protein localization to kinetochore / SUMOylation of RNA binding proteins / structural constituent of nuclear pore / SUMOylation of chromatin organization proteins / RNA export from nucleus / silent mating-type cassette heterochromatin formation / nucleocytoplasmic transport / vacuolar membrane / poly(A)+ mRNA export from nucleus / nuclear localization sequence binding / regulation of mitotic nuclear division / dynein intermediate chain binding / NLS-bearing protein import into nucleus / cytoplasmic microtubule / establishment of mitotic spindle orientation / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; セリンエンドペプチターゼ / ribosomal large subunit export from nucleus / positive regulation of TOR signaling / subtelomeric heterochromatin formation / mRNA transport / mRNA export from nucleus / nuclear pore / ribosomal small subunit export from nucleus / ERAD pathway / Neutrophil degranulation / positive regulation of TORC1 signaling / protein export from nucleus / cellular response to amino acid starvation / nuclear periphery / cell periphery / chromosome segregation / promoter-specific chromatin binding / molecular condensate scaffold activity / heterochromatin formation / phospholipid binding / protein import into nucleus / transcription corepressor activity / double-strand break repair / protein transport / nuclear envelope / single-stranded DNA binding / cellular response to heat / nuclear membrane / amyloid fibril formation / chromosome, telomeric region / hydrolase activity / cell division / chromatin binding / endoplasmic reticulum membrane / protein-containing complex binding / positive regulation of DNA-templated transcription / structural molecule activity / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / positive regulation of transcription by RNA polymerase II / DNA binding / RNA binding / identical protein binding / nucleus / cytosol / cytoplasm 類似検索 - 分子機能 | ||||||||||||||||||||||||
| 生物種 |  | ||||||||||||||||||||||||
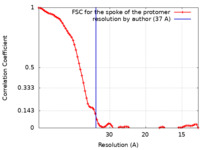
| 手法 | サブトモグラム平均法 / クライオ電子顕微鏡法 / 解像度: 37.0 Å | ||||||||||||||||||||||||
 データ登録者 データ登録者 | Villa E / Singh D | ||||||||||||||||||||||||
| 資金援助 |  米国, 7件 米国, 7件
| ||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Cell / 年: 2022 ジャーナル: Cell / 年: 2022タイトル: Comprehensive structure and functional adaptations of the yeast nuclear pore complex. 著者: Christopher W Akey / Digvijay Singh / Christna Ouch / Ignacia Echeverria / Ilona Nudelman / Joseph M Varberg / Zulin Yu / Fei Fang / Yi Shi / Junjie Wang / Daniel Salzberg / Kangkang Song / ...著者: Christopher W Akey / Digvijay Singh / Christna Ouch / Ignacia Echeverria / Ilona Nudelman / Joseph M Varberg / Zulin Yu / Fei Fang / Yi Shi / Junjie Wang / Daniel Salzberg / Kangkang Song / Chen Xu / James C Gumbart / Sergey Suslov / Jay Unruh / Sue L Jaspersen / Brian T Chait / Andrej Sali / Javier Fernandez-Martinez / Steven J Ludtke / Elizabeth Villa / Michael P Rout /  要旨: Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub- ...Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub-nanometer resolution to show how flexible connectors tie together different structural and functional layers. These connectors may be targets for phosphorylation and regulated disassembly in cells with an open mitosis. Moreover, some nucleoporin pairs and transport factors have similar interaction motifs, which suggests an evolutionary and mechanistic link between assembly and transport. We provide evidence for three major NPC variants that may foreshadow functional specializations at the nuclear periphery. Cryo-electron tomography extended these studies, providing a model of the in situ NPC with a radially expanded inner ring. Our comprehensive model reveals features of the nuclear basket and central transporter, suggests a role for the lumenal Pom152 ring in restricting dilation, and highlights structural plasticity that may be required for transport. | ||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_24258.map.gz emd_24258.map.gz | 76.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-24258-v30.xml emd-24258-v30.xml emd-24258.xml emd-24258.xml | 68.6 KB 68.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_24258_fsc.xml emd_24258_fsc.xml | 7.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_24258.png emd_24258.png | 197.2 KB | ||
| マスクデータ |  emd_24258_msk_1.map emd_24258_msk_1.map emd_24258_msk_2.map emd_24258_msk_2.map emd_24258_msk_3.map emd_24258_msk_3.map | 34.3 MB 34.3 MB 34.3 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-24258.cif.gz emd-24258.cif.gz | 18.9 KB | ||
| その他 |  emd_24258_additional_1.map.gz emd_24258_additional_1.map.gz emd_24258_additional_2.map.gz emd_24258_additional_2.map.gz emd_24258_additional_3.map.gz emd_24258_additional_3.map.gz emd_24258_additional_4.map.gz emd_24258_additional_4.map.gz emd_24258_additional_5.map.gz emd_24258_additional_5.map.gz emd_24258_additional_6.map.gz emd_24258_additional_6.map.gz emd_24258_additional_7.map.gz emd_24258_additional_7.map.gz emd_24258_half_map_1.map.gz emd_24258_half_map_1.map.gz emd_24258_half_map_2.map.gz emd_24258_half_map_2.map.gz | 111 MB 111.7 MB 113.8 MB 111.9 MB 111.8 MB 111.2 MB 112.2 MB 12.7 MB 12.8 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24258 http://ftp.pdbj.org/pub/emdb/structures/EMD-24258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24258 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_24258_validation.pdf.gz emd_24258_validation.pdf.gz | 935 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_24258_full_validation.pdf.gz emd_24258_full_validation.pdf.gz | 934.6 KB | 表示 | |
| XML形式データ |  emd_24258_validation.xml.gz emd_24258_validation.xml.gz | 16.2 KB | 表示 | |
| CIF形式データ |  emd_24258_validation.cif.gz emd_24258_validation.cif.gz | 20.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24258 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24258 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24258 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24258 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7n9fMC  7n84C  7n85C C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_24258.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_24258.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | full in situ NPC map recombined from 90 degree wedge focused alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 6.74 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
+マスク #1
+マスク #2
+マスク #3
+追加マップ: zoned 3D map of Nsp1-FG-connections
+追加マップ: zoned 3D map of inner ring of spokes
+追加マップ: full in situ NPC map recombined from 90...
+追加マップ: zoned 3D map of the N-ring
+追加マップ: zoned 3D map of C-ring plus Nup82 complex with 3 Dyn 2 dimers
+追加マップ: zoned 3D map of Pom152 ring
+追加マップ: difference map for pore membrane
+ハーフマップ: half map
+ハーフマップ: half map
- 試料の構成要素
試料の構成要素
+全体 : yeast NPC
+超分子 #1: yeast NPC
+分子 #1: Nucleoporin NUP170
+分子 #2: Nucleoporin NUP157
+分子 #3: orphans bound to Nup192 NTD
+分子 #4: Nucleoporin NSP1
+分子 #5: Nucleoporin NUP57
+分子 #6: Nucleoporin NUP49/NSP49
+分子 #7: Nucleoporin NUP192
+分子 #8: Nucleoporin NUP188
+分子 #9: Nucleoporin NIC96
+分子 #10: Nucleoporin NUP53
+分子 #11: Nucleoporin ASM4
+分子 #12: Nucleoporin NUP82
+分子 #13: Nucleoporin NUP159
+分子 #14: Nucleoporin NUP120
+分子 #15: Nucleoporin NUP85
+分子 #16: Nucleoporin 145c
+分子 #17: Protein transport protein SEC13
+分子 #18: Nucleoporin SEH1
+分子 #19: Nucleoporin NUP84
+分子 #20: Nucleoporin NUP133
+分子 #21: Dynein light chain 1, cytoplasmic
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | サブトモグラム平均法 |
| 試料の集合状態 | cell |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER / メッシュ: 200 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 60 sec. / 前処理 - 気圧: 0.039 kPa |
| 凍結 | 凍結剤: ETHANE-PROPANE 詳細: A custom-built vitrification device (Max Planck Institute for Biochemistry, Munich). |
| 詳細 | W303 yeast cells were harvested during log-phase growth and diluted to an 0.8 x 107 cells/mL in YPD media. Five uL of this diluted sample was applied to glow-discharged 200-mesh, Quantafoil R2/1 grids (Electron Microscopy Sciences), excess media was manually blotted from the grid back side (opposite to the carbon substrate where the cells were deposited) and the grid was plunge frozen in a liquid ethane-propane mixture (50/50 volume, Airgas) then cryogenic, FIB milling was performed in an Aquilos DualBeam (Thermo Fisher Scientific) |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Quantum LS / エネルギーフィルター - スリット幅: 20 eV |
| 詳細 | Tilt series collection: dose-symmetric & bi-directional |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 3838 pixel / デジタル化 - サイズ - 縦: 3708 pixel / 平均露光時間: 2.0 sec. / 平均電子線量: 4.5 e/Å2 詳細: Images were collected in movie-mode at ~8 frames per second |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 最大 デフォーカス(補正後): 6.0 µm / 最小 デフォーカス(補正後): 2.5 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 6.0 µm / 最小 デフォーカス(公称値): 2.5 µm / 倍率(公称値): 42000 |
| 試料ステージ | ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 詳細 | Manual docking and limited molecular dynamics flexible fitting |
|---|---|
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT / 当てはまり具合の基準: cross-correlation |
| 得られたモデル |  PDB-7n9f: |
 ムービー
ムービー コントローラー
コントローラー



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)