[English] 日本語
 Yorodumi
Yorodumi- EMDB-24095: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24095 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | |||||||||
 Map data Map data | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Anaplastic lymphoma kinase / Receptor tyrosine kinases / RTK / FAM150 / DE NOVO PROTEIN / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of ERK5 cascade / ASP-3026-resistant ALK mutants / NVP-TAE684-resistant ALK mutants / alectinib-resistant ALK mutants / brigatinib-resistant ALK mutants / ceritinib-resistant ALK mutants / crizotinib-resistant ALK mutants / lorlatinib-resistant ALK mutants / MDK and PTN in ALK signaling / receptor signaling protein tyrosine kinase activator activity ...positive regulation of ERK5 cascade / ASP-3026-resistant ALK mutants / NVP-TAE684-resistant ALK mutants / alectinib-resistant ALK mutants / brigatinib-resistant ALK mutants / ceritinib-resistant ALK mutants / crizotinib-resistant ALK mutants / lorlatinib-resistant ALK mutants / MDK and PTN in ALK signaling / receptor signaling protein tyrosine kinase activator activity / regulation of dopamine receptor signaling pathway / response to environmental enrichment / transmembrane receptor protein tyrosine kinase activator activity / ALK mutants bind TKIs / phosphorylation / Signaling by LTK / positive regulation of dendrite development / regulation of neuron differentiation / peptidyl-tyrosine autophosphorylation / Signaling by ALK / adult behavior / response to stress / negative regulation of lipid catabolic process / neuron development / energy homeostasis / swimming behavior / transmembrane receptor protein tyrosine kinase activity / cell surface receptor protein tyrosine kinase signaling pathway / cytokine activity / hippocampus development / positive regulation of neuron projection development / receptor protein-tyrosine kinase / : / receptor tyrosine kinase binding / Signaling by ALK fusions and activated point mutants / regulation of cell population proliferation / heparin binding / protein autophosphorylation / protein tyrosine kinase activity / regulation of apoptotic process / positive regulation of ERK1 and ERK2 cascade / receptor complex / signal transduction / protein-containing complex / extracellular space / extracellular exosome / extracellular region / ATP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
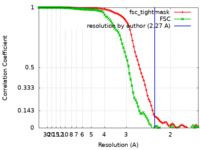
| Method | single particle reconstruction / cryo EM / Resolution: 2.27 Å | |||||||||
 Authors Authors | Myasnikov AG / Reshetnyak AV / Kalodimos CG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Mechanism for the activation of the anaplastic lymphoma kinase receptor. Authors: Andrey V Reshetnyak / Paolo Rossi / Alexander G Myasnikov / Munia Sowaileh / Jyotidarsini Mohanty / Amanda Nourse / Darcie J Miller / Irit Lax / Joseph Schlessinger / Charalampos G Kalodimos /  Abstract: Anaplastic lymphoma kinase (ALK) is a receptor tyrosine kinase (RTK) that regulates important functions in the central nervous system. The ALK gene is a hotspot for chromosomal translocation events ...Anaplastic lymphoma kinase (ALK) is a receptor tyrosine kinase (RTK) that regulates important functions in the central nervous system. The ALK gene is a hotspot for chromosomal translocation events that result in several fusion proteins that cause a variety of human malignancies. Somatic and germline gain-of-function mutations in ALK were identified in paediatric neuroblastoma. ALK is composed of an extracellular region (ECR), a single transmembrane helix and an intracellular tyrosine kinase domain. ALK is activated by the binding of ALKAL1 and ALKAL2 ligands to its ECR, but the lack of structural information for the ALK-ECR or for ALKAL ligands has limited our understanding of ALK activation. Here we used cryo-electron microscopy, nuclear magnetic resonance and X-ray crystallography to determine the atomic details of human ALK dimerization and activation by ALKAL1 and ALKAL2. Our data reveal a mechanism of RTK activation that allows dimerization by either dimeric (ALKAL2) or monomeric (ALKAL1) ligands. This mechanism is underpinned by an unusual architecture of the receptor-ligand complex. The ALK-ECR undergoes a pronounced ligand-induced rearrangement and adopts an orientation parallel to the membrane surface. This orientation is further stabilized by an interaction between the ligand and the membrane. Our findings highlight the diversity in RTK oligomerization and activation mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24095.map.gz emd_24095.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24095-v30.xml emd-24095-v30.xml emd-24095.xml emd-24095.xml | 31.5 KB 31.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24095_fsc.xml emd_24095_fsc.xml emd_24095_fsc_2.xml emd_24095_fsc_2.xml | 9.7 KB 9.1 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_24095.png emd_24095.png | 74.8 KB | ||
| Filedesc metadata |  emd-24095.cif.gz emd-24095.cif.gz | 7.3 KB | ||
| Others |  emd_24095_additional_1.map.gz emd_24095_additional_1.map.gz emd_24095_additional_2.map.gz emd_24095_additional_2.map.gz emd_24095_additional_3.map.gz emd_24095_additional_3.map.gz emd_24095_half_map_1.map.gz emd_24095_half_map_1.map.gz emd_24095_half_map_2.map.gz emd_24095_half_map_2.map.gz | 59.9 MB 32.1 MB 5 MB 48.6 MB 48.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24095 http://ftp.pdbj.org/pub/emdb/structures/EMD-24095 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24095 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24095 | HTTPS FTP |
-Related structure data
| Related structure data |  7n00MC  7mzwC  7mzxC  7mzyC  7mzzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10930 (Title: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alpha EMPIAR-10930 (Title: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alphaData size: 10.2 TB Data #1: Unaligned multi-frame micrographs of 2:2 ALK:AugAlpha complex [micrographs - multiframe] Data #2: Aligned single-frame micrographs of 2:2 ALK:AugAlpha complex [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24095.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24095.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
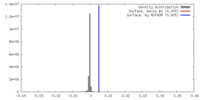
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand...
| File | emd_24095_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | ||||||||||||
| Projections & Slices |
| ||||||||||||
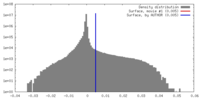
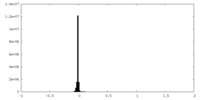
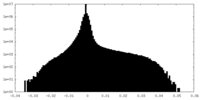
| Density Histograms |
-Additional map: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand...
| File | emd_24095_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | ||||||||||||
| Projections & Slices |
| ||||||||||||
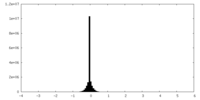
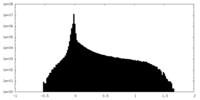
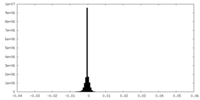
| Density Histograms |
-Additional map: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand...
| File | emd_24095_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | ||||||||||||
| Projections & Slices |
| ||||||||||||
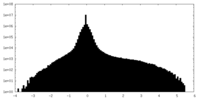
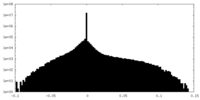
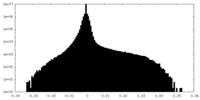
| Density Histograms |
-Half map: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand...
| File | emd_24095_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | ||||||||||||
| Projections & Slices |
| ||||||||||||
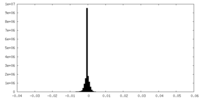
| Density Histograms |
-Half map: Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand...
| File | emd_24095_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-1025 in complex with AUG-alpha | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : hetero-tetrameric complex of anaplastic lymphoma kinase (ALK) wit...
| Entire | Name: hetero-tetrameric complex of anaplastic lymphoma kinase (ALK) with Augmentor alpha |
|---|---|
| Components |
|
-Supramolecule #1: hetero-tetrameric complex of anaplastic lymphoma kinase (ALK) wit...
| Supramolecule | Name: hetero-tetrameric complex of anaplastic lymphoma kinase (ALK) with Augmentor alpha type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 108 kDa/nm |
-Macromolecule #1: ALK tyrosine kinase receptor
| Macromolecule | Name: ALK tyrosine kinase receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.377559 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GTAPKSRNLF ERNPNKELKP GENSPRQTPI FDPTVHWLFT TCGASGPHGP TQAQCNNAYQ NSNLSVEVGS EGPLKGIQIW KVPATDTYS ISGYGAAGGK GGKNTMMRSH GVSVLGIFNL EKDDMLYILV GQQGEDACPS TNQLIQKVCI GENNVIEEEI R VNRSVHEW ...String: GTAPKSRNLF ERNPNKELKP GENSPRQTPI FDPTVHWLFT TCGASGPHGP TQAQCNNAYQ NSNLSVEVGS EGPLKGIQIW KVPATDTYS ISGYGAAGGK GGKNTMMRSH GVSVLGIFNL EKDDMLYILV GQQGEDACPS TNQLIQKVCI GENNVIEEEI R VNRSVHEW AGGGGGGGGA TYVFKMKDGV PVPLIIAAGG GGRAYGAKTD TFHPERLENN SSVLGLNGNS GAAGGGGGWN DN TSLLWAG KSLQEGATGG HSCPQAMKKW GWETRGGFGG GGGGCSSGGG GGGYIGGNAA SNNDPEMDGE DGVSFISPLG ILY TPALKV MEGHGEVNIK HYLNCSHCEV DECHMDPESH KVICFCDHGT VLAEDGVSCI VSP UniProtKB: ALK tyrosine kinase receptor |
-Macromolecule #2: ALK and LTK ligand 2
| Macromolecule | Name: ALK and LTK ligand 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.640013 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAEPREPADG QALLRLVVEL VQELRKHHSA EHKGLQLLGR DYALGRAEAA GLGPSPEQRV EIVPRDLRMK DKFLKHLTGP LYFSPKCSK HFHRLYHNTR DCTIPAYYKR CARLLTRLAV SPVCMEDKQ UniProtKB: ALK and LTK ligand 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: QUANTIFOIL R1.2/1/3 gold 300 mesh grids without glow-discharge, blotting time 3 sec., blotting force -5.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 96.0 K / Max: 98.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV / Details: Gatan energy filter |
| Details | SerialEM coma-free alignment |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 2498 / Average exposure time: 4.2 sec. / Average electron dose: 81.0 e/Å2 / Details: 4.2 second exposure, 70 frames, total dose 81e |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7n00: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)