[English] 日本語
 Yorodumi
Yorodumi- EMDB-23698: Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Parti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23698 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
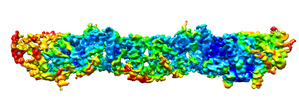
| Title | Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle Cryo-EM | ||||||||||||
 Map data Map data | Human septin hexameric complex SEPT2G-SEPT6-SEPT7. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Complex / SPA / Cytoskeleton / CELL CYCLE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationseptin collar / regulation of embryonic cell shape / sperm annulus / positive regulation of non-motile cilium assembly / septin complex / photoreceptor connecting cilium / septin ring / cytoskeleton-dependent cytokinesis / regulation of exocytosis / non-motile cilium ...septin collar / regulation of embryonic cell shape / sperm annulus / positive regulation of non-motile cilium assembly / septin complex / photoreceptor connecting cilium / septin ring / cytoskeleton-dependent cytokinesis / regulation of exocytosis / non-motile cilium / ciliary membrane / smoothened signaling pathway / intercellular bridge / cell division site / cleavage furrow / axoneme / mitotic cytokinesis / cilium assembly / stress fiber / axon terminus / Anchoring of the basal body to the plasma membrane / MAPK6/MAPK4 signaling / kinetochore / spindle / microtubule cytoskeleton / protein localization / actin cytoskeleton / synaptic vesicle / midbody / spermatogenesis / molecular adaptor activity / cell differentiation / cadherin binding / GTPase activity / GTP binding / structural molecule activity / extracellular exosome / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
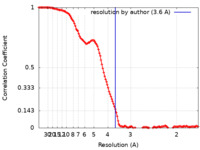
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Mendonca DC / Pereira HM | ||||||||||||
| Funding support |  Brazil, 3 items Brazil, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2021 Journal: J Mol Biol / Year: 2021Title: An atomic model for the human septin hexamer by cryo-EM. Authors: Deborah C Mendonça / Samuel L Guimarães / Humberto D'Muniz Pereira / Andressa A Pinto / Marcelo A de Farias / Andre S de Godoy / Ana P U Araujo / Marin van Heel / Rodrigo V Portugal / Richard C Garratt /  Abstract: In order to form functional filaments, human septins must assemble into hetero-oligomeric rod-like particles which polymerize end-to-end. The rules governing the assembly of these particles and the ...In order to form functional filaments, human septins must assemble into hetero-oligomeric rod-like particles which polymerize end-to-end. The rules governing the assembly of these particles and the subsequent filaments are incompletely understood. Although crystallographic approaches have been successful in studying the separate components of the system, there has been difficulty in obtaining high resolution structures of the full particle. Here we report a first cryo-EM structure for a hexameric rod composed of human septins 2, 6 and 7 with a global resolution of ~3.6 Å and a local resolution of between ~3.0 Å and ~5.0 Å. By fitting the previously determined high-resolution crystal structures of the component subunits into the cryo-EM map, we are able to provide an essentially complete model for the particle. This exposes SEPT2 NC-interfaces at the termini of the hexamer and leaves internal cavities between the SEPT6-SEPT7 pairs. The floor of the cavity is formed by the two α helices including their polybasic regions. These are locked into place between the two subunits by interactions made with the α and α helices of the neighbouring monomer together with its polyacidic region. The cavity may serve to provide space allowing the subunits to move with respect to one another. The elongated particle shows a tendency to bend at its centre where two copies of SEPT7 form a homodimeric G-interface. Such bending is almost certainly related to the ability of septin filaments to recognize and even induce membrane curvature. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23698.map.gz emd_23698.map.gz | 450.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23698-v30.xml emd-23698-v30.xml emd-23698.xml emd-23698.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23698_fsc.xml emd_23698_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23698.png emd_23698.png | 393.9 KB | ||
| Filedesc metadata |  emd-23698.cif.gz emd-23698.cif.gz | 5.9 KB | ||
| Others |  emd_23698_half_map_1.map.gz emd_23698_half_map_1.map.gz emd_23698_half_map_2.map.gz emd_23698_half_map_2.map.gz | 442.8 MB 442.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23698 http://ftp.pdbj.org/pub/emdb/structures/EMD-23698 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23698 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23698 | HTTPS FTP |
-Validation report
| Summary document |  emd_23698_validation.pdf.gz emd_23698_validation.pdf.gz | 901 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23698_full_validation.pdf.gz emd_23698_full_validation.pdf.gz | 900.5 KB | Display | |
| Data in XML |  emd_23698_validation.xml.gz emd_23698_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_23698_validation.cif.gz emd_23698_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23698 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23698 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23698 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23698 | HTTPS FTP |
-Related structure data
| Related structure data |  7m6jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23698.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23698.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human septin hexameric complex SEPT2G-SEPT6-SEPT7. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle...
| File | emd_23698_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle Cryo-EM | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle...
| File | emd_23698_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle Cryo-EM | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7
| Entire | Name: Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 |
|---|---|
| Components |
|
-Supramolecule #1: Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7
| Supramolecule | Name: Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Septin-2
| Macromolecule | Name: Septin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.748352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGFEFTLMVV GESGLGKSTL INSLFLTDLY PERVIPGAAE KIERTVQIEA STVEIEERGV KLRLTVVDTP GYGDAINCRD CFKTIISYI DEQFERYLHD ESGLNRRHII DNRVHCCFYF ISPFGHGLKP LDVAFMKAIH NKVNIVPVIA KADTLTLKER E RLKKRILD ...String: MGFEFTLMVV GESGLGKSTL INSLFLTDLY PERVIPGAAE KIERTVQIEA STVEIEERGV KLRLTVVDTP GYGDAINCRD CFKTIISYI DEQFERYLHD ESGLNRRHII DNRVHCCFYF ISPFGHGLKP LDVAFMKAIH NKVNIVPVIA KADTLTLKER E RLKKRILD EIEEHNIKIY HLPDAESDED EDFKEQTRLL KASIPFSVVG SNQLIEAKGK KVRGRLYPWG VVEVENPEHN DF LKLRTML ITHMQDLQEV TQDLHYENFR SERLKRGG UniProtKB: Septin-2 |
-Macromolecule #2: Septin-6
| Macromolecule | Name: Septin-6 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.948723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAATDIARQV GEGCRTVPLA GHVGFDSLPD QLVNKSVSQG FCFNILCVGE TGLGKSTLMD TLFNTKFEGE PATHTQPGVQ LQSNTYDLQ ESNVRLKLTI VSTVGFGDQI NKEDSYKPIV EFIDAQFEAY LQEELKIRRV LHTYHDSRIH VCLYFIAPTG H SLKSLDLV ...String: MAATDIARQV GEGCRTVPLA GHVGFDSLPD QLVNKSVSQG FCFNILCVGE TGLGKSTLMD TLFNTKFEGE PATHTQPGVQ LQSNTYDLQ ESNVRLKLTI VSTVGFGDQI NKEDSYKPIV EFIDAQFEAY LQEELKIRRV LHTYHDSRIH VCLYFIAPTG H SLKSLDLV TMKKLDSKVN IIPIIAKADA ISKSELTKFK IKITSELVSN GVQIYQFPTD DESVAEINGT MNAHLPFAVI GS TEELKIG NKMMRARQYP WGTVQVENEA HCDFVKLREM LIRVNMEDLR EQTHTRHYEL YRRCKLEEMG FKDTDPDSKP FSL QETYEA KRNEFLGELQ KKEEEMRQMF VQRVKEKEAE LKEAEKELHE KFDRLKKLHQ DEKKKLEDKK KSLDDEVNAF KQRK TAAEL LQSQGSQAGG SQTLKRDKEK KN UniProtKB: Septin-6 |
-Macromolecule #3: Septin-7
| Macromolecule | Name: Septin-7 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.456645 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPMVAQQK NLEGYVGFAN LPNQVYRKSV KRGFEFTLMV VGESGLGKST LINSLFLTDL YSPEYPGPSH RIKKTVQVE QSKVLIKEGG VQLLLTIVDT PGFGDAVDNS NCWQPVIDYI DSKFEDYLNA ESRVNRRQMP DNRVQCCLYF I APSGHGLK ...String: MGSSHHHHHH SQDPMVAQQK NLEGYVGFAN LPNQVYRKSV KRGFEFTLMV VGESGLGKST LINSLFLTDL YSPEYPGPSH RIKKTVQVE QSKVLIKEGG VQLLLTIVDT PGFGDAVDNS NCWQPVIDYI DSKFEDYLNA ESRVNRRQMP DNRVQCCLYF I APSGHGLK PLDIEFMKRL HEKVNIIPLI AKADTLTPEE CQQFKKQIMK EIQEHKIKIY EFPETDDEEE NKLVKKIKDR LP LAVVGSN TIIEVNGKRV RGRQYPWGVA EVENGEHCDF TILRNMLIRT HMQDLKDVTN NVHYENYRSR KLAAVTYNGV DNN KNKGQL TKSPLAQMEE ERREHVAKMK KMEMEMEQVF EMKVKEKVQK LKDSEAELQR RHEQMKKNLE AQHKELEEKR RQFE DEKAN WEAQQRILEQ QNSSRTLEKN KKKGKIF UniProtKB: Septin-7 |
-Macromolecule #4: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)