+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23570 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of yeast DNA Polymerase Zeta (apo) | |||||||||
 Map data Map data | cryo-em map of apo-PolZ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleic acid binding / DNA polymerase / metal ion binding / catalytic activity / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdelta DNA polymerase complex / H3-H4 histone complex chaperone activity / DNA amplification / zeta DNA polymerase complex / RNA-templated DNA biosynthetic process / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI ...delta DNA polymerase complex / H3-H4 histone complex chaperone activity / DNA amplification / zeta DNA polymerase complex / RNA-templated DNA biosynthetic process / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / DNA replication, removal of RNA primer / lagging strand elongation / double-strand break repair via break-induced replication / DNA damage tolerance / error-free translesion synthesis / DNA metabolic process / DNA strand elongation involved in DNA replication / leading strand elongation / error-prone translesion synthesis / mismatch repair / nucleotide-excision repair / double-strand break repair via homologous recombination / base-excision repair / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / chromatin / mitochondrion / DNA binding / zinc ion binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.11 Å | |||||||||
 Authors Authors | Truong CD / Craig TA | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2021 Journal: J Biol Chem / Year: 2021Title: Cryo-EM reveals conformational flexibility in apo DNA polymerase ζ. Authors: Chloe Du Truong / Theodore A Craig / Gaofeng Cui / Maria Victoria Botuyan / Rachel A Serkasevich / Ka-Yi Chan / Georges Mer / Po-Lin Chiu / Rajiv Kumar /  Abstract: The translesion synthesis (TLS) DNA polymerases Rev1 and Polζ function together in DNA lesion bypass during DNA replication, acting as nucleotide inserter and extender polymerases, respectively. ...The translesion synthesis (TLS) DNA polymerases Rev1 and Polζ function together in DNA lesion bypass during DNA replication, acting as nucleotide inserter and extender polymerases, respectively. While the structural characterization of the Saccharomyces cerevisiae Polζ in its DNA-bound state has illuminated how this enzyme synthesizes DNA, a mechanistic understanding of TLS also requires probing conformational changes associated with DNA- and Rev1 binding. Here, we used single-particle cryo-electron microscopy to determine the structure of the apo Polζ holoenzyme. We show that compared with its DNA-bound state, apo Polζ displays enhanced flexibility that correlates with concerted motions associated with expansion of the Polζ DNA-binding channel upon DNA binding. We also identified a lysine residue that obstructs the DNA-binding channel in apo Polζ, suggesting a gating mechanism. The Polζ subunit Rev7 is a hub protein that directly binds Rev1 and is a component of several other protein complexes such as the shieldin DNA double-strand break repair complex. We analyzed the molecular interactions of budding yeast Rev7 in the context of Polζ and those of human Rev7 in the context of shieldin using a crystal structure of Rev7 bound to a fragment of the shieldin-3 protein. Overall, our study provides new insights into Polζ mechanism of action and the manner in which Rev7 recognizes partner proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23570.map.gz emd_23570.map.gz | 37.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23570-v30.xml emd-23570-v30.xml emd-23570.xml emd-23570.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23570.png emd_23570.png | 53.9 KB | ||
| Filedesc metadata |  emd-23570.cif.gz emd-23570.cif.gz | 8.3 KB | ||
| Others |  emd_23570_additional_1.map.gz emd_23570_additional_1.map.gz emd_23570_additional_2.map.gz emd_23570_additional_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23570 http://ftp.pdbj.org/pub/emdb/structures/EMD-23570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23570 | HTTPS FTP |
-Validation report
| Summary document |  emd_23570_validation.pdf.gz emd_23570_validation.pdf.gz | 554.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23570_full_validation.pdf.gz emd_23570_full_validation.pdf.gz | 553.8 KB | Display | |
| Data in XML |  emd_23570_validation.xml.gz emd_23570_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_23570_validation.cif.gz emd_23570_validation.cif.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23570 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23570 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23570 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23570 | HTTPS FTP |
-Related structure data
| Related structure data |  7lxdMC  6ve5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23570.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23570.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-em map of apo-PolZ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.491 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
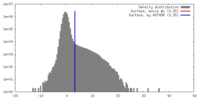
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Half-map 1
| File | emd_23570_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
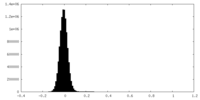
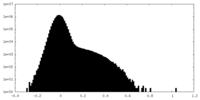
| Density Histograms |
-Additional map: Half-map 2
| File | emd_23570_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
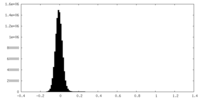
| Density Histograms |
- Sample components
Sample components
-Entire : DNA polymerase Zeta
| Entire | Name: DNA polymerase Zeta |
|---|---|
| Components |
|
-Supramolecule #1: DNA polymerase Zeta
| Supramolecule | Name: DNA polymerase Zeta / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: DNA polymerase Zeta is generated from yeast without DNA binding. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 172 kDa/nm |
-Macromolecule #1: DNA polymerase zeta catalytic subunit
| Macromolecule | Name: DNA polymerase zeta catalytic subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 173.197531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRESNDTIQ SDTVRSSSKS DYFRIQLNNQ DYYMSKPTFL DPSHGESLPL NQFSQVPNIR VFGALPTGHQ VLCHVHGILP YMFIKYDGQ ITDTSTLRHQ RCAQVHKTLE VKIRASFKRK KDDKHDLAGD KLGNLNFVAD VSVVKGIPFY GYHVGWNLFY K ISLLNPSC ...String: MSRESNDTIQ SDTVRSSSKS DYFRIQLNNQ DYYMSKPTFL DPSHGESLPL NQFSQVPNIR VFGALPTGHQ VLCHVHGILP YMFIKYDGQ ITDTSTLRHQ RCAQVHKTLE VKIRASFKRK KDDKHDLAGD KLGNLNFVAD VSVVKGIPFY GYHVGWNLFY K ISLLNPSC LSRISELIRD GKIFGKKFEI YESHIPYLLQ WTADFNLFGC SWINVDRCYF RSPVLNSILD IDKLTINDDL QL LLDRFCD FKCNVLSRRD FPRVGNGLIE IDILPQFIKN REKLQHRDIH HDFLEKLGDI SDIPVKPYVS SARDMINELT MQR EELSLK EYKEPPETKR HVSGHQWQSS GEFEAFYKKA QHKTSTFDGQ IPNFENFIDK NQKFSAINTP YEALPQLWPR LPQI EINNN SMQDKKNDDQ VNASFTEYEI CGVDNENEGV KGSNIKSRSY SWLPESIASP KDSTILLDHQ TKYHNTINFS MDCAM TQNM ASKRKLRSSV SANKTSLLSR KRKKVMAAGL RYGKRAFVYG EPPFGYQDIL NKLEDEGFPK IDYKDPFFSN PVDLEN KPY AYAGKRFEIS STHVSTRIPV QFGGETVSVY NKPTFDMFSS WKYALKPPTY DAVQKWYNKV PSMGNKKTES QISMHTP HS KFLYKFASDV SGKQKRKKSS VHDSLTHLTL EIHANTRSDK IPDPAIDEVS MIIWCLEEET FPLDLDIAYE GIMIVHKA S EDSTFPTKIQ HCINEIPVMF YESEFEMFEA LTDLVLLLDP DILSGFEIHN FSWGYIIERC QKIHQFDIVR ELARVKCQI KTKLSDTWGY AHSSGIMITG RHMINIWRAL RSDVNLTQYT IESAAFNILH KRLPHFSFES LTNMWNAKKS TTELKTVLNY WLSRAQINI QLLRKQDYIA RNIEQARLIG IDFHSVYYRG SQFKVESFLI RICKSESFIL LSPGKKDVRK QKALECVPLV M EPESAFYK SPLIVLDFQS LYPSIMIGYN YCYSTMIGRV REINLTENNL GVSKFSLPRN ILALLKNDVT IAPNGVVYAK TS VRKSTLS KMLTDILDVR VMIKKTMNEI GDDNTTLKRL LNNKQLALKL LANVTYGYTS ASFSGRMPCS DLADSIVQTG RET LEKAID IIEKDETWNA KVVYGDTDSL FVYLPGKTAI EAFSIGHAMA ERVTQNNPKP IFLKFEKVYH PSILISKKRY VGFS YESPS QTLPIFDAKG IETVRRDGIP AQQKIIEKCI RLLFQTKDLS KIKKYLQNEF FKIQIGKVSA QDFCFAKEVK LGAYK SEKT APAGAVVVKR RINEDHRAEP QYKERIPYLV VKGKQGQLLR ERCVSPEEFL EGENLELDSE YYINKILIPP LDRLFN LIG INVGNWAQEI VKSKRASTTT TKVENITRVG TSATCCNCGE ELTKICSLQL CDDCLEKRST TTLSFLIKKL KRQKEYQ TL KTVCRTCSYR YTSDAGIEND HIASKCNSYD CPVFYSRVKA ERYLRDNQSV QREEALISLN DW UniProtKB: DNA polymerase zeta catalytic subunit |
-Macromolecule #2: DNA polymerase zeta processivity subunit
| Macromolecule | Name: DNA polymerase zeta processivity subunit / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 28.791654 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNRWVEKWLR VYLKCYINLI LFYRNVYPPQ SFDYTTYQSF NLPQFVPINR HPALIDYIEE LILDVLSKLT HVYRFSICII NKKNDLCIE KYVLDFSELQ HVDKDDQIIT ETEVFDEFRS SLNSLIMHLE KLPKVNDDTI TFEAVINAIE LELGHKLDRN R RVDSLEEK ...String: MNRWVEKWLR VYLKCYINLI LFYRNVYPPQ SFDYTTYQSF NLPQFVPINR HPALIDYIEE LILDVLSKLT HVYRFSICII NKKNDLCIE KYVLDFSELQ HVDKDDQIIT ETEVFDEFRS SLNSLIMHLE KLPKVNDDTI TFEAVINAIE LELGHKLDRN R RVDSLEEK AEIERDSNWV KCQEDENLPD NNGFQPPKIK LTSLVGSDVG PLIIHQFSEK LISGDDKILN GVYSQYEEGE SI FGSLF UniProtKB: DNA polymerase zeta processivity subunit |
-Macromolecule #3: DNA polymerase delta small subunit
| Macromolecule | Name: DNA polymerase delta small subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 55.603992 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LHMDALLTKF NEDRSLQDEN LSQPRTRVRI VDDNLYNKSN PFQLCYKKRD YGSQYYHIYQ YRLKTFRERV LKECDKRWDA GFTLNGQLV LKKDKVLDIQ GNQPCWCVGS IYCEMKYKPN VLDEVINDTY GAPDLTKSYT DKEGGSDEIM LEDESGRVLL V GDFIRSTP ...String: LHMDALLTKF NEDRSLQDEN LSQPRTRVRI VDDNLYNKSN PFQLCYKKRD YGSQYYHIYQ YRLKTFRERV LKECDKRWDA GFTLNGQLV LKKDKVLDIQ GNQPCWCVGS IYCEMKYKPN VLDEVINDTY GAPDLTKSYT DKEGGSDEIM LEDESGRVLL V GDFIRSTP FITGVVVGIL GMEAEAGTFQ VLDICYPTPL PQNPFPAPIA TCPTRGKIAL VSGLNLNNTS PDRLLRLEIL RE FLMGRIN NKIDDISLIG RLLICGNSVD FDIKSVNKDE LMISLTEFSK FLHNILPSIS VDIMPGTNDP SDKSLPQQPF HKS LFDKSL ESYFNGSNKE ILNLVTNPYE FSYNGVDVLA VSGKNINDIC KYVIPSNDNG ESENKVEEGE SNDFKDDIEH RLDL MECTM KWQNIAPTAP DTLWCYPYTD KDPFVLDKWP HVYIVANQPY FGTRVVEIGG KNIKIISVPE FSSTGMIILL DLETL EAET VKIDI UniProtKB: DNA polymerase delta small subunit |
-Macromolecule #4: DNA polymerase delta subunit 3
| Macromolecule | Name: DNA polymerase delta subunit 3 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 40.377715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDQKASYFIN EKLFTEVKPV LFTDLIHHLK IGPSMAKKLM FDYYKQTTNA KYNCVVICCY KDQTIKIIHD LSNIPQQDSI IDCFIYAFN PMDSFIPYYD IIDQKDCLTI KNSYELKVSE SSKIIERTKT LEEKSKPLVR PTARSKTTPE ETTGRKSKSK D MGLRSTAL ...String: MDQKASYFIN EKLFTEVKPV LFTDLIHHLK IGPSMAKKLM FDYYKQTTNA KYNCVVICCY KDQTIKIIHD LSNIPQQDSI IDCFIYAFN PMDSFIPYYD IIDQKDCLTI KNSYELKVSE SSKIIERTKT LEEKSKPLVR PTARSKTTPE ETTGRKSKSK D MGLRSTAL LAKMKKDRDD KETSRQNELR KRKEENLQKI NKQNPEREAQ MKELNNLFVE DDLDTEEVNG GSKPNSPKET DS NDKDKNN DDLEDLLETT AEDSLMDVPK IQQTKPSETE HSKEPKSEEE PSSFIDEDGY IVTKRPATST PPRKPSPVVK RAL SSSKKQ ETPSSNKRLK KQGTLESFFK RKAK UniProtKB: DNA polymerase delta subunit 3 |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.9 Component:
Details: Solutions were made fresh from concentrate to avoid microbial contamination. They are further filtered using 0.2 micrometer filtering membrane and a pressure/vacuum filtration unit. | |||||||||||||||
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK II / Details: Blot for 6 seconds before plunging. | |||||||||||||||
| Details | This sample was mono disperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Number real images: 11698 / Average exposure time: 6.0 sec. / Average electron dose: 45.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 48780 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)