[English] 日本語
 Yorodumi
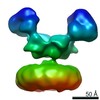
Yorodumi- EMDB-23562: Negative stain EM map of 1E01 Fab in complex with N2 Singapore16 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23562 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of 1E01 Fab in complex with N2 Singapore16 | |||||||||
 Map data Map data | Negative stain EM map of 1E01 Fab in complex with N2 Singapore16 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information: / : / : / exo-alpha-sialidase / carbohydrate metabolic process / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 14.9 Å | |||||||||
 Authors Authors | Turner HL / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Antibodies targeting the neuraminidase active site inhibit influenza H3N2 viruses with an S245N glycosylation site. Authors: Daniel Stadlbauer / Meagan McMahon / Hannah L Turner / Xueyong Zhu / Hongquan Wan / Juan Manuel Carreño / George O'Dell / Shirin Strohmeier / Zain Khalil / Marta Luksza / Harm van Bakel / ...Authors: Daniel Stadlbauer / Meagan McMahon / Hannah L Turner / Xueyong Zhu / Hongquan Wan / Juan Manuel Carreño / George O'Dell / Shirin Strohmeier / Zain Khalil / Marta Luksza / Harm van Bakel / Viviana Simon / Ali H Ellebedy / Ian A Wilson / Andrew B Ward / Florian Krammer /   Abstract: Contemporary influenza A H3N2 viruses circulating since 2016 have acquired a glycosylation site in the neuraminidase in close proximity to the enzymatic active site. Here, we investigate if this ...Contemporary influenza A H3N2 viruses circulating since 2016 have acquired a glycosylation site in the neuraminidase in close proximity to the enzymatic active site. Here, we investigate if this S245N glycosylation site, as a result of antigenic evolution, can impact binding and function of human monoclonal antibodies that target the conserved active site. While we find that a reduction in the inhibitory ability of neuraminidase active site binders is measurable, this class of broadly reactive monoclonal antibodies maintains protective efficacy in vivo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23562.map.gz emd_23562.map.gz | 8.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23562-v30.xml emd-23562-v30.xml emd-23562.xml emd-23562.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23562.png emd_23562.png | 84.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23562 http://ftp.pdbj.org/pub/emdb/structures/EMD-23562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23562 | HTTPS FTP |
-Validation report
| Summary document |  emd_23562_validation.pdf.gz emd_23562_validation.pdf.gz | 327.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23562_full_validation.pdf.gz emd_23562_full_validation.pdf.gz | 327.2 KB | Display | |
| Data in XML |  emd_23562_validation.xml.gz emd_23562_validation.xml.gz | 5.6 KB | Display | |
| Data in CIF |  emd_23562_validation.cif.gz emd_23562_validation.cif.gz | 6.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23562 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23562 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23562.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23562.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM map of 1E01 Fab in complex with N2 Singapore16 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.77 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 1E01 antibody-Neuraminidase N2 Singapore16 complex
| Entire | Name: 1E01 antibody-Neuraminidase N2 Singapore16 complex |
|---|---|
| Components |
|
-Supramolecule #1: 1E01 antibody-Neuraminidase N2 Singapore16 complex
| Supramolecule | Name: 1E01 antibody-Neuraminidase N2 Singapore16 complex / type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #3: 1E01 antibody
| Supramolecule | Name: 1E01 antibody / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Supramolecule #2: Neuraminidase N2 Singapore16
| Supramolecule | Name: Neuraminidase N2 Singapore16 / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl formate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 14.9 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 40555 |
|---|---|
| Initial angle assignment | Type: OTHER / Software - Name: RELION (ver. 3.0) / Details: bayesian polishing |
| Final angle assignment | Type: OTHER / Software - Name: RELION (ver. 3.0) / Details: bayesian poilishing |
| Final 3D classification | Software - Name: RELION (ver. 3.0) |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















