[English] 日本語
 Yorodumi
Yorodumi- EMDB-2310: Three-dimensional structure of active, full-length human telomera... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2310 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional structure of active, full-length human telomerase dimer, determined by single-particle electron microscopy in negative stain | |||||||||
 Map data Map data | 3D map of human telomerase dimer bound to G-overhang oligonucleotide (TTAGGG)2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Telomerase reverse transcriptase / human / telomere length extension | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation / RNA-directed RNA polymerase complex / siRNA transcription / positive regulation of protein localization to nucleolus / telomerase catalytic core complex / RNA-templated DNA biosynthetic process ...positive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation / RNA-directed RNA polymerase complex / siRNA transcription / positive regulation of protein localization to nucleolus / telomerase catalytic core complex / RNA-templated DNA biosynthetic process / establishment of protein localization to telomere / telomerase activity / Regulation of MITF-M-dependent genes involved in DNA replication, damage repair and senescence / nuclear telomere cap complex / siRNA processing / telomere maintenance via recombination / positive regulation of vascular associated smooth muscle cell migration / telomerase holoenzyme complex / telomerase RNA binding / DNA biosynthetic process / RNA-templated transcription / telomeric DNA binding / positive regulation of stem cell proliferation / mitochondrial nucleoid / negative regulation of cellular senescence / replicative senescence / Telomere Extension By Telomerase / positive regulation of Wnt signaling pathway / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of G1/S transition of mitotic cell cycle / response to cadmium ion / positive regulation of protein binding / telomere maintenance via telomerase / negative regulation of endothelial cell apoptotic process / positive regulation of vascular associated smooth muscle cell proliferation / telomere maintenance / positive regulation of D-glucose import / mitochondrion organization / Formation of the beta-catenin:TCF transactivating complex / PML body / regulation of protein stability / positive regulation of miRNA transcription / RNA-directed DNA polymerase / transcription coactivator binding / RNA-directed DNA polymerase activity / positive regulation of angiogenesis / protein import into nucleus / protein-folding chaperone binding / heart development / cellular response to hypoxia / negative regulation of neuron apoptotic process / tRNA binding / chromosome, telomeric region / nuclear speck / RNA-directed RNA polymerase activity / nucleolus / protein homodimerization activity / DNA binding / RNA binding / nucleoplasm / metal ion binding / identical protein binding / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Sauerwald A / Sandin S / Cristofari G / Scheres SHW / Lingner J / Rhodes D | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2013 Journal: Nat Struct Mol Biol / Year: 2013Title: Structure of active dimeric human telomerase. Authors: Anselm Sauerwald / Sara Sandin / Gaël Cristofari / Sjors H W Scheres / Joachim Lingner / Daniela Rhodes /  Abstract: Telomerase contains a large RNA subunit, TER, and a protein catalytic subunit, TERT. Whether telomerase functions as a monomer or dimer has been a matter of debate. Here we report biochemical and ...Telomerase contains a large RNA subunit, TER, and a protein catalytic subunit, TERT. Whether telomerase functions as a monomer or dimer has been a matter of debate. Here we report biochemical and labeling data that show that in vivo-assembled human telomerase contains two TERT subunits and binds two telomeric DNA substrates. Notably, catalytic activity requires both TERT active sites to be functional, which demonstrates that human telomerase functions as a dimer. We also present the three-dimensional structure of the active full-length human telomerase dimer, determined by single-particle EM in negative stain. Telomerase has a bilobal architecture with the two monomers linked by a flexible interface. The monomer reconstruction at 23-Å resolution and fitting of the atomic structure of the TERT subunit from beetle Tribolium castaneum into the EM density reveals the spatial relationship between RNA and protein subunits, providing insights into telomerase architecture. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2310.map.gz emd_2310.map.gz | 133.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2310-v30.xml emd-2310-v30.xml emd-2310.xml emd-2310.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2310.png EMD-2310.png | 39.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2310 http://ftp.pdbj.org/pub/emdb/structures/EMD-2310 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2310 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2310 | HTTPS FTP |
-Validation report
| Summary document |  emd_2310_validation.pdf.gz emd_2310_validation.pdf.gz | 188.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2310_full_validation.pdf.gz emd_2310_full_validation.pdf.gz | 187.9 KB | Display | |
| Data in XML |  emd_2310_validation.xml.gz emd_2310_validation.xml.gz | 4.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2310 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2310 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2310 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2310 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2310.map.gz / Format: CCP4 / Size: 144.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2310.map.gz / Format: CCP4 / Size: 144.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of human telomerase dimer bound to G-overhang oligonucleotide (TTAGGG)2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Three-dimensional structure of active, full-length human telomera...
| Entire | Name: Three-dimensional structure of active, full-length human telomerase dimer, determined by single-particle electron microscopy in negative stain |
|---|---|
| Components |
|
-Supramolecule #1000: Three-dimensional structure of active, full-length human telomera...
| Supramolecule | Name: Three-dimensional structure of active, full-length human telomerase dimer, determined by single-particle electron microscopy in negative stain type: sample / ID: 1000 Details: The composition analysed by mass spectroscopy.Purified telomerase contains the hTERT subunits and two accessory proteins, Nop10 and dyskerin. Telomerase complexes have a molecular weight ...Details: The composition analysed by mass spectroscopy.Purified telomerase contains the hTERT subunits and two accessory proteins, Nop10 and dyskerin. Telomerase complexes have a molecular weight consistent with that of a dimer consisting of two hTERT (127 kDa) and two hTER (153 kDa) subunits, as well as the two accessory proteins Nop10 (7.7 kDa) and dyskerin (58 kDA). Oligomeric state: Dimer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 670 KDa / Theoretical: 626 KDa / Method: Sedimentation |
-Macromolecule #1: Telomerase reverse transcriptase
| Macromolecule | Name: Telomerase reverse transcriptase / type: protein_or_peptide / ID: 1 / Name.synonym: TERT / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: Human Embryonic Kidney cells Homo sapiens (human) / synonym: Human / Cell: Human Embryonic Kidney cells |
| Molecular weight | Experimental: 127 KDa / Theoretical: 670 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK239T / Recombinant plasmid: pCDNA6 Homo sapiens (human) / Recombinant cell: HEK239T / Recombinant plasmid: pCDNA6 |
| Sequence | UniProtKB: Telomerase reverse transcriptase / GO: telomere maintenance / InterPro: Telomerase reverse transcriptase |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 20 mM Tris, 150 mM KCl, 1 mM MgCl2 |
| Staining | Type: NEGATIVE Details: Continuous carbon-coated grids were freshly prepared and glow-discharged before use. 13 microl of telomerase sample (8-10 nM) were deposited on the grid for 15-30 minutes, blotted with ...Details: Continuous carbon-coated grids were freshly prepared and glow-discharged before use. 13 microl of telomerase sample (8-10 nM) were deposited on the grid for 15-30 minutes, blotted with filter paper and negatively stained with 2 drops of 1-2% (w/v) uranyl acetate solution. |
| Grid | Details: 200 mesh carbon coated with thin carbon, glow discharged |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Corrected at 100,000 times magnification |
| Details | The films were developed in Kodak developer at full strength for 12 min. |
| Date | Jan 1, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 482 / Average electron dose: 10 e/Å2 / Details: The micrographs were compressed x4 / Od range: 1.4 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Calibrated magnification: 42550 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.0015 µm / Nominal defocus min: 0.001 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Details | Electron tomography was used to calculate a low-resolution reference model. Distinct conformations were further classified by three-dimensional maximum-likelihood analysis in Fourier space (MLF3D).Density map calculated from a sub-population of 3,659 particles. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: EMAN2, XMIPP Details: 3D structure was refined using the low-resolution subtomogram average as a reference model for 3D single particle refinement with EMAN2. Distinct conformations were further classified by ...Details: 3D structure was refined using the low-resolution subtomogram average as a reference model for 3D single particle refinement with EMAN2. Distinct conformations were further classified by three-dimensional maximum-likelihood analysis in Fourier space (MLF3D). Density map calculated from a sub-population of 3,659 particles. The absolute hand and the overall correctness of the dimer structure were assessed using a modified version of tilt-pair validation method. Number images used: 20127 |
| Final two d classification | Number classes: 100 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera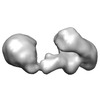










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















