+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3055 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The vacuolar H+-ATPase masked around Vo | |||||||||
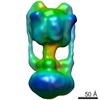
 Map data Map data | Reconstruction of the Vo domain from the V-ATPase using masking | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rotary ATPase / vacuolar ATPase | |||||||||
| Biological species |  Manduca sexta (tobacco hornworm) Manduca sexta (tobacco hornworm) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.8 Å | |||||||||
 Authors Authors | Rawson S / Tiburcy F / Trinick J / Wieczorek H / Harrison MA / Muench SP | |||||||||
 Citation Citation |  Journal: Methods / Year: 2016 Journal: Methods / Year: 2016Title: Methods to account for movement and flexibility in cryo-EM data processing. Authors: S Rawson / M G Iadanza / N A Ranson / S P Muench /  Abstract: Recent advances in direct electron detectors and improved CMOS cameras have been accompanied by the development of a range of software to take advantage of the data they produce. In particular they ...Recent advances in direct electron detectors and improved CMOS cameras have been accompanied by the development of a range of software to take advantage of the data they produce. In particular they allow for the correction of two types of motion in cryo electron microscopy samples: motion correction for movements of the sample particles in the ice, and differential masking to account for heterogeneity caused by flexibility within protein complexes. Here we provide several scripts that allow users to move between RELION and standalone motion correction and centring programs. We then compare the computational cost and improvements in data quality with each program. We also describe our masking procedures to account for conformational flexibility. For the different elements of this study we have used three samples; a high symmetry virus, flexible protein complex (∼1MDa) and a relatively small protein complex (∼550kDa), to benchmark four widely available motion correction packages. Using these as test cases we demonstrate how motion correction and differential masking, as well as an additional particle re-centring protocol can improve final reconstructions when used within the RELION image-processing package. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3055.map.gz emd_3055.map.gz | 10 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3055-v30.xml emd-3055-v30.xml emd-3055.xml emd-3055.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  Vo_13499_3055.tiff Vo_13499_3055.tiff | 252.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3055 http://ftp.pdbj.org/pub/emdb/structures/EMD-3055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3055 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3055.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3055.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the Vo domain from the V-ATPase using masking | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Manduca sexta vacuolar ATPase complex
| Entire | Name: Manduca sexta vacuolar ATPase complex |
|---|---|
| Components |
|
-Supramolecule #1000: Manduca sexta vacuolar ATPase complex
| Supramolecule | Name: Manduca sexta vacuolar ATPase complex / type: sample / ID: 1000 / Details: monodisperse / Oligomeric state: monomeric / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 900 KDa / Method: mass spec |
-Supramolecule #1: Vacuolar ATPase
| Supramolecule | Name: Vacuolar ATPase / type: organelle_or_cellular_component / ID: 1 / Name.synonym: V-ATPase / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Manduca sexta (tobacco hornworm) / synonym: tobacco hornworm Manduca sexta (tobacco hornworm) / synonym: tobacco hornworm |
| Molecular weight | Experimental: 900 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL |
|---|---|
| Buffer | pH: 8.1 Details: 150 mM NaCl, 20 mM Tris-HCl, 9.6 mM 2-mercaptoethanol, 0.01% C12E10 |
| Grid | Details: 400 mesh Quantifoil R2.0/2.0 grids with thin carbon (10 nm) coating, glow discharged in air. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK IV / Method: Grids were blotted for 7.5 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Collected with FEI EPU software |
| Date | Feb 27, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 1366 / Average electron dose: 60 e/Å2 Details: Each micrograph is sum of 34 frames recorded by direct detector. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 103704 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Standard procedures in RELION1.3, using mask around Vo |
|---|---|
| CTF correction | Details: Relion |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.8 Å / Resolution method: OTHER / Software - Name: Relion Details: Maximum likelihood in Relion using 3D auto-refine and a mask around Vo. The particles were handpicked in BOXER Number images used: 13083 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















