[English] 日本語
 Yorodumi
Yorodumi- EMDB-22918: Tetrameric human mitochondrial Hsp90 (TRAP1) in the presence of A... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22918 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetrameric human mitochondrial Hsp90 (TRAP1) in the presence of AMP-PNP | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Hsp90 / CHAPERONE / TRAP1 / SdhB / mitochondria / SpyTag / SpyCatcher | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtranslational attenuation / negative regulation of cellular respiration / Oxidoreductases; Acting on the CH-OH group of donors; With a quinone or similar compound as acceptor / succinate metabolic process / respiratory chain complex II (succinate dehydrogenase) / mitochondrial electron transport, succinate to ubiquinone / Citric acid cycle (TCA cycle) / succinate dehydrogenase (quinone) activity / succinate dehydrogenase / Maturation of TCA enzymes and regulation of TCA cycle ...translational attenuation / negative regulation of cellular respiration / Oxidoreductases; Acting on the CH-OH group of donors; With a quinone or similar compound as acceptor / succinate metabolic process / respiratory chain complex II (succinate dehydrogenase) / mitochondrial electron transport, succinate to ubiquinone / Citric acid cycle (TCA cycle) / succinate dehydrogenase (quinone) activity / succinate dehydrogenase / Maturation of TCA enzymes and regulation of TCA cycle / Respiratory electron transport / tumor necrosis factor receptor binding / 3 iron, 4 sulfur cluster binding / negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide / : / ubiquinone binding / proton motive force-driven mitochondrial ATP synthesis / negative regulation of reactive oxygen species biosynthetic process / tricarboxylic acid cycle / aerobic respiration / respiratory electron transport chain / ATP-dependent protein folding chaperone / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / unfolded protein binding / protein folding / 4 iron, 4 sulfur cluster binding / electron transfer activity / mitochondrial inner membrane / mitochondrial matrix / protein kinase binding / ATP hydrolysis activity / mitochondrion / RNA binding / nucleoplasm / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||||||||||||||
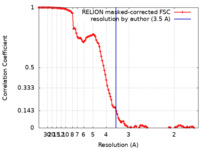
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||||||||
 Authors Authors | Liu YX / Agard DA | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM analysis of human mitochondrial Hsp90 in multiple tetrameric states Authors: Liu YX / Agard DA / Elnatan D / Sun M / Larson AG | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22918.map.gz emd_22918.map.gz | 116.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22918-v30.xml emd-22918-v30.xml emd-22918.xml emd-22918.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22918_fsc.xml emd_22918_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22918.png emd_22918.png | 166.9 KB | ||
| Filedesc metadata |  emd-22918.cif.gz emd-22918.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22918 http://ftp.pdbj.org/pub/emdb/structures/EMD-22918 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22918 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22918 | HTTPS FTP |
-Related structure data
| Related structure data |  7kluMC  7klvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22918.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22918.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8488 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tetrameric human mitochondrial Hsp90 (TRAP1) in the presence of A...
| Entire | Name: Tetrameric human mitochondrial Hsp90 (TRAP1) in the presence of AMP-PNP |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric human mitochondrial Hsp90 (TRAP1) in the presence of A...
| Supramolecule | Name: Tetrameric human mitochondrial Hsp90 (TRAP1) in the presence of AMP-PNP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Heat shock protein 75 kDa, mitochondrial, SpyCatcher
| Macromolecule | Name: Heat shock protein 75 kDa, mitochondrial, SpyCatcher / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 87.094617 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIDPFTSTQT AEDKEEPLHS IISSTESVQG STSKHEFQAE TKKLLDIVAR SLYSEKEVFI RELISNASDA LEKLRHKLVS DGQALPEME IHLQTNAEKG TITIQDTGIG MTQEELVSNL GTIARSGSKA FLDALQNQAE ASSKIIGQFG VGFYSAFMVA D RVEVYSRS ...String: GIDPFTSTQT AEDKEEPLHS IISSTESVQG STSKHEFQAE TKKLLDIVAR SLYSEKEVFI RELISNASDA LEKLRHKLVS DGQALPEME IHLQTNAEKG TITIQDTGIG MTQEELVSNL GTIARSGSKA FLDALQNQAE ASSKIIGQFG VGFYSAFMVA D RVEVYSRS AAPGSLGYQW LSDGSGVFEI AEASGVRTGT KIIIHLKSDC KEFSSEARVR DVVTKYSNFV SFPLYLNGRR MN TLQAIWM MDPKDVGEWQ HEEFYRYVAQ AHDKPRYTLH YKTDAPLNIR SIFYVPDMKP SMFDVSRELG SSVALYSRKV LIQ TKATDI LPKWLRFIRG VVDSEDIPLN LSRELLQESA LIRKLRDVLQ QRLIKFFIDQ SKKDAEKYAK FFEDYGLFMR EGIV TATEQ EVKEDIAKLL RYESSALPSG QLTSLSEYAS RMRAGTRNIY YLCAPNRHLA EHSPYYEAMK KKDTEVLFCF EQFDE LTLL HLREFDKKKL ISVETDIVVD HYKEEKFEDR SPAAECLSEK ETEELMAWMR NVLGSRVTNV KVTLRLDTHP AMVTVL EMG AARHFLRMQQ LAKTQEERAQ LLQPTLEINP RHALIKKLNQ LRASEPGLAQ LLVDQIYENA MIAAGLVDDP RAMVGRL NE LLVKALERHG GSGSGSSAMV DTLSGLSSEQ GQSGDMTIEE DSATHIKFSK RDEDGKELAG ATMELRDSSG KTISTWIS D GQVKDFYLYP GKYTFVETAA PDGYEVATAI TFTVNEQGQV TVNGKATKGD AHI UniProtKB: Heat shock protein 75 kDa, mitochondrial |
-Macromolecule #2: Heat shock protein 75 kDa, mitochondrial, SpyTag, Succinate dehyd...
| Macromolecule | Name: Heat shock protein 75 kDa, mitochondrial, SpyTag, Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial (chimera) type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: succinate dehydrogenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.121344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIDPFTSTQT AEDKEEPLHS IISSTESVQG STSKHEFQAE TKKLLDIVAR SLYSEKEVFI RELISNASDA LEKLRHKLVS DGQALPEME IHLQTNAEKG TITIQDTGIG MTQEELVSNL GTIARSGSKA FLDALQNQAE ASSKIIGQFG VGFYSAFMVA D RVEVYSRS ...String: GIDPFTSTQT AEDKEEPLHS IISSTESVQG STSKHEFQAE TKKLLDIVAR SLYSEKEVFI RELISNASDA LEKLRHKLVS DGQALPEME IHLQTNAEKG TITIQDTGIG MTQEELVSNL GTIARSGSKA FLDALQNQAE ASSKIIGQFG VGFYSAFMVA D RVEVYSRS AAPGSLGYQW LSDGSGVFEI AEASGVRTGT KIIIHLKSDC KEFSSEARVR DVVTKYSNFV SFPLYLNGRR MN TLQAIWM MDPKDVGEWQ HEEFYRYVAQ AHDKPRYTLH YKTDAPLNIR SIFYVPDMKP SMFDVSRELG SSVALYSRKV LIQ TKATDI LPKWLRFIRG VVDSEDIPLN LSRELLQESA LIRKLRDVLQ QRLIKFFIDQ SKKDAEKYAK FFEDYGLFMR EGIV TATEQ EVKEDIAKLL RYESSALPSG QLTSLSEYAS RMRAGTRNIY YLCAPNRHLA EHSPYYEAMK KKDTEVLFCF EQFDE LTLL HLREFDKKKL ISVETDIVVD HYKEEKFEDR SPAAECLSEK ETEELMAWMR NVLGSRVTNV KVTLRLDTHP AMVTVL EMG AARHFLRMQQ LAKTQEERAQ LLQPTLEINP RHALIKKLNQ LRASEPGLAQ LLVDQIYENA MIAAGLVDDP RAMVGRL NE LLVKALERHG GSGSGSSAHI VMVDAYKPTK GGGGSGGGGS GGGGSLEVLF QGPGSAQTAA ATAPRIKKFA IYRWDPDK A GDKPHMQTYE VDLNKCGPMV LDALIKIKNE VDSTLTFRRS CREGICGSCA MNINGGNTLA CTRRIDTNLN KVSKIYPLP HMYVIKDLVP DLSNFYAQYK SIEPYLKKK UniProtKB: Heat shock protein 75 kDa, mitochondrial, Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 4 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)