[English] 日本語
 Yorodumi
Yorodumi- EMDB-22876: Cryo-EM Structure of the Desulfovibrio vulgaris Type I-C Apo Cascade -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22876 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Desulfovibrio vulgaris Type I-C Apo Cascade | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / Cascade / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / hydrolase activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) / Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) /  Desulfovibrio vulgaris str. Hildenborough (bacteria) Desulfovibrio vulgaris str. Hildenborough (bacteria) | |||||||||
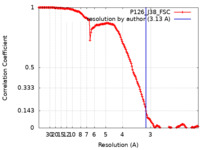
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | O'Brien R / Wrapp D / Bravo JPK / Schwartz E / Taylor D | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structural basis for assembly of non-canonical small subunits into type I-C Cascade. Authors: Roisin E O'Brien / Inês C Santos / Daniel Wrapp / Jack P K Bravo / Evan A Schwartz / Jennifer S Brodbelt / David W Taylor /  Abstract: Bacteria and archaea employ CRISPR (clustered, regularly, interspaced, short palindromic repeats)-Cas (CRISPR-associated) systems as a type of adaptive immunity to target and degrade foreign nucleic ...Bacteria and archaea employ CRISPR (clustered, regularly, interspaced, short palindromic repeats)-Cas (CRISPR-associated) systems as a type of adaptive immunity to target and degrade foreign nucleic acids. While a myriad of CRISPR-Cas systems have been identified to date, type I-C is one of the most commonly found subtypes in nature. Interestingly, the type I-C system employs a minimal Cascade effector complex, which encodes only three unique subunits in its operon. Here, we present a 3.1 Å resolution cryo-EM structure of the Desulfovibrio vulgaris type I-C Cascade, revealing the molecular mechanisms that underlie RNA-directed complex assembly. We demonstrate how this minimal Cascade utilizes previously overlooked, non-canonical small subunits to stabilize R-loop formation. Furthermore, we describe putative PAM and Cas3 binding sites. These findings provide the structural basis for harnessing the type I-C Cascade as a genome-engineering tool. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22876.map.gz emd_22876.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22876-v30.xml emd-22876-v30.xml emd-22876.xml emd-22876.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22876_fsc.xml emd_22876_fsc.xml | 16.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22876.png emd_22876.png | 163.8 KB | ||
| Filedesc metadata |  emd-22876.cif.gz emd-22876.cif.gz | 6.7 KB | ||
| Others |  emd_22876_half_map_1.map.gz emd_22876_half_map_1.map.gz emd_22876_half_map_2.map.gz emd_22876_half_map_2.map.gz | 285.3 MB 285.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22876 http://ftp.pdbj.org/pub/emdb/structures/EMD-22876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22876 | HTTPS FTP |
-Related structure data
| Related structure data |  7khaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22876.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22876.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_22876_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_22876_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Type I-C Apo Cascade
| Entire | Name: Type I-C Apo Cascade |
|---|---|
| Components |
|
-Supramolecule #1: Type I-C Apo Cascade
| Supramolecule | Name: Type I-C Apo Cascade / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Minimal CRISPR Cascade |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria)Strain: Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303 |
| Molecular weight | Theoretical: 371 KDa |
-Macromolecule #1: CRISPR-associated protein, CT1134 family
| Macromolecule | Name: CRISPR-associated protein, CT1134 family / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria)Strain: Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303 |
| Molecular weight | Theoretical: 25.250969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TYGIRLRVWG DYACFTRPEM KVERVSYDVM PPSAARGILE AIHWKPAIRW IVDRIHVLRP IVFDNVRRNE VSSKIPKPNP ATAMRDRKP LYFLVDDGSN RQQRAATLLR NVDYVIEAHF ELTDKAGAED NAGKHLDIFR RRARAGQSFQ QPCLGCREFP A SFELLEGD ...String: TYGIRLRVWG DYACFTRPEM KVERVSYDVM PPSAARGILE AIHWKPAIRW IVDRIHVLRP IVFDNVRRNE VSSKIPKPNP ATAMRDRKP LYFLVDDGSN RQQRAATLLR NVDYVIEAHF ELTDKAGAED NAGKHLDIFR RRARAGQSFQ QPCLGCREFP A SFELLEGD VPLSCYAGEK RDLGYMLLDI DFERDMTPLF FKAVMEDGVI TPPSRTSPEV RA UniProtKB: pre-crRNA processing endonuclease |
-Macromolecule #2: CRISPR-associated protein, TM1801 family
| Macromolecule | Name: CRISPR-associated protein, TM1801 family / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria)Strain: Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303 |
| Molecular weight | Theoretical: 32.358912 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTAIANRYEF VLLFDVENGN PNGDPDAGNM PRIDPETGHG LVTDVCLKRK IRNHVALTKE GAERFNIYIQ EKAILNETHE RAYTACDLK PEPKKLPKKV EDAKRVTDWM CTNFYDIRTF GAVMTTEVNC GQVRGPVQMA FARSVEPVVP QEVSITRMAV T TKAEAEKQ ...String: MTAIANRYEF VLLFDVENGN PNGDPDAGNM PRIDPETGHG LVTDVCLKRK IRNHVALTKE GAERFNIYIQ EKAILNETHE RAYTACDLK PEPKKLPKKV EDAKRVTDWM CTNFYDIRTF GAVMTTEVNC GQVRGPVQMA FARSVEPVVP QEVSITRMAV T TKAEAEKQ QGDNRTMGRK HIVPYGLYVA HGFISAPLAE KTGFSDEDLT LFWDALVNMF EHDRSAARGL MSSRKLIVFK HQ NRLGNAP AHKLFDLVKV SRAEGSSGPA RSFADYAVTV GQAPEGVEVK EML UniProtKB: CRISPR-associated protein, TM1801 family |
-Macromolecule #3: CRISPR-associated protein, CT1133 family
| Macromolecule | Name: CRISPR-associated protein, CT1133 family / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria)Strain: Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303 |
| Molecular weight | Theoretical: 59.310883 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: WENTSYILGV DAKGKQERTD KCHAAFIAHI KAYCDTADQD LAAVLQFLEH GEKDLSAFPV SEEVIGSNIV FRIEGEPGFV HERPAARQA WANCLNRREQ GLCGQCLITG ERQKPIAQLH PSIKGGRDGV RGAQAVASIV SFNNTAFESY GKEQSINAPV S QEAAFSYV ...String: WENTSYILGV DAKGKQERTD KCHAAFIAHI KAYCDTADQD LAAVLQFLEH GEKDLSAFPV SEEVIGSNIV FRIEGEPGFV HERPAARQA WANCLNRREQ GLCGQCLITG ERQKPIAQLH PSIKGGRDGV RGAQAVASIV SFNNTAFESY GKEQSINAPV S QEAAFSYV TALNYLLNPS NRQKVTIADA TVVFWAERSS PAEDIFAGMF DPPSTTAKPE SSNGTPPEDS EEGSQPDTAR DD PHAAARM HDLLVAIRSG KRATDIMPDM DESVRFHVLG LSPNAARLSV RFWEVDTVGH MLDKVGRHYR ELEIIPQFNN EQE FPSLST LLRQTAVLNK TENISPVLAG GLFRAMLTGG PYPQSLLPAV LGRIRAEHAR PEDKSRYRLE VVTYYRAALI KAYL IRNRK LEVPVSLDPA RTDRPYLLGR LFAVLEKAQE DAVPGANATI KDRYLASASA NPGQVFHMLL KNASNHTAKL RKDPE RKGS AIHYEIMMQE IIDNISDFPV TMSSDEQGLF MIGYYHQRKA LFTKKNKEN UniProtKB: CRISPR-associated protein, CT1133 family |
-Macromolecule #4: CRISPR-associated protein, CT1133 family
| Macromolecule | Name: CRISPR-associated protein, CT1133 family / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303) (bacteria)Strain: Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303 |
| Molecular weight | Theoretical: 14.017981 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VSLDPARTDR PYLLGRLFAV LEKAQEDAVP GANATIKDRY LASASANPGQ VFHMLLKNAS NHTAKLRKDP ERKGSAIHYE IMMQEIIDN ISDFPVTMSS DEQGLFMIGY YHQRKALFTK KNKEN UniProtKB: CRISPR-associated protein, CT1133 family |
-Macromolecule #5: RNA (45-MER)
| Macromolecule | Name: RNA (45-MER) / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris str. Hildenborough (bacteria) Desulfovibrio vulgaris str. Hildenborough (bacteria)Strain: Hildenborough / ATCC 29579 / DSM 644 / NCIMB 8303 |
| Molecular weight | Theoretical: 14.457629 KDa |
| Sequence | String: UGGAUUGAAA CGCCAUGCUC AGGCUGGCGA GUGCGCCACU CAUCA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-2/2 / Material: COPPER / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 5400 / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)