[English] 日本語
 Yorodumi
Yorodumi- EMDB-2279: Electron cryo-microscopy of a head-tailed virus infecting extreme... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2279 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of a head-tailed virus infecting extremely halophilic archaea | |||||||||
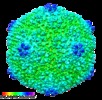
 Map data Map data | Reconstruction of HSTV-1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Icosahedral capsid | |||||||||
| Biological species | HSTV-1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.9 Å | |||||||||
 Authors Authors | Pietila MK / Laurinmaki P / Russell DA / Ko CC / Jacobs-Sera D / Butcher SJ / Bamford DH / Hendrix RW | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2013 Journal: Proc Natl Acad Sci U S A / Year: 2013Title: Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Authors: Maija K Pietilä / Pasi Laurinmäki / Daniel A Russell / Ching-Chung Ko / Deborah Jacobs-Sera / Roger W Hendrix / Dennis H Bamford / Sarah J Butcher /  Abstract: It has been proposed that viruses can be divided into a small number of structure-based viral lineages. One of these lineages is exemplified by bacterial virus Hong Kong 97 (HK97), which represents ...It has been proposed that viruses can be divided into a small number of structure-based viral lineages. One of these lineages is exemplified by bacterial virus Hong Kong 97 (HK97), which represents the head-tailed dsDNA bacteriophages. Seemingly similar viruses also infect archaea. Here we demonstrate using genomic analysis, electron cryomicroscopy, and image reconstruction that the major coat protein fold of newly isolated archaeal Haloarcula sinaiiensis tailed virus 1 has the canonical coat protein fold of HK97. Although it has been anticipated previously, this is physical evidence that bacterial and archaeal head-tailed viruses share a common architectural principle. The HK97-like fold has previously been recognized also in herpesviruses, and this study expands the HK97-like lineage to viruses from all three domains of life. This is only the second established lineage to include archaeal, bacterial, and eukaryotic viruses. Thus, our findings support the hypothesis that the last common universal ancestor of cellular organisms was infected by a number of different viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2279.map.gz emd_2279.map.gz | 71.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2279-v30.xml emd-2279-v30.xml emd-2279.xml emd-2279.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2279.tif emd_2279.tif | 961.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2279 http://ftp.pdbj.org/pub/emdb/structures/EMD-2279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2279 | HTTPS FTP |
-Validation report
| Summary document |  emd_2279_validation.pdf.gz emd_2279_validation.pdf.gz | 285 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2279_full_validation.pdf.gz emd_2279_full_validation.pdf.gz | 284.1 KB | Display | |
| Data in XML |  emd_2279_validation.xml.gz emd_2279_validation.xml.gz | 6.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2279 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2279 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2279.map.gz / Format: CCP4 / Size: 278 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2279.map.gz / Format: CCP4 / Size: 278 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of HSTV-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
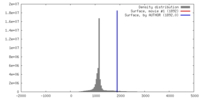
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : archaeal virus HSTV-1
| Entire | Name: archaeal virus HSTV-1 |
|---|---|
| Components |
|
-Supramolecule #1000: archaeal virus HSTV-1
| Supramolecule | Name: archaeal virus HSTV-1 / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: HSTV-1
| Supramolecule | Name: HSTV-1 / type: virus / ID: 1 / Sci species name: HSTV-1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Haloarcula sinaiiensis ATCC 33800 (Halophile) / synonym: ARCHAEA Haloarcula sinaiiensis ATCC 33800 (Halophile) / synonym: ARCHAEA |
| Virus shell | Shell ID: 1 / Diameter: 624 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Details: 20mM Tris-HCl |
|---|---|
| Grid | Details: Quantifoil holey carbon film on 300 mesh copper grid. |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Sep 9, 2009 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Bits/pixel: 12 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 62000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 8.9 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: AUTO3DEM / Number images used: 7115 |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)