[English] 日本語
 Yorodumi
Yorodumi- EMDB-22678: Structure of apo VCP hexamer generated from bacterially recombina... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22678 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of apo VCP hexamer generated from bacterially recombinant VCP/p97 | |||||||||
 Map data Map data | Cryo-EM density map of apo VCP hexamer generated from bacterially recombinant VCP/p97. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA ATPase / ATP hydrolysis / segregase / CYTOSOLIC PROTEIN / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology information: / flavin adenine dinucleotide catabolic process / VCP-NSFL1C complex / endosome to lysosome transport via multivesicular body sorting pathway / endoplasmic reticulum stress-induced pre-emptive quality control / BAT3 complex binding / cellular response to arsenite ion / protein-DNA covalent cross-linking repair / Derlin-1 retrotranslocation complex / positive regulation of protein K63-linked deubiquitination ...: / flavin adenine dinucleotide catabolic process / VCP-NSFL1C complex / endosome to lysosome transport via multivesicular body sorting pathway / endoplasmic reticulum stress-induced pre-emptive quality control / BAT3 complex binding / cellular response to arsenite ion / protein-DNA covalent cross-linking repair / Derlin-1 retrotranslocation complex / positive regulation of protein K63-linked deubiquitination / cytoplasm protein quality control / positive regulation of oxidative phosphorylation / : / aggresome assembly / deubiquitinase activator activity / mitotic spindle disassembly / ubiquitin-modified protein reader activity / regulation of protein localization to chromatin / VCP-NPL4-UFD1 AAA ATPase complex / cellular response to misfolded protein / negative regulation of protein localization to chromatin / positive regulation of mitochondrial membrane potential / vesicle-fusing ATPase / K48-linked polyubiquitin modification-dependent protein binding / regulation of aerobic respiration / retrograde protein transport, ER to cytosol / stress granule disassembly / ATPase complex / regulation of synapse organization / ubiquitin-specific protease binding / positive regulation of ATP biosynthetic process / MHC class I protein binding / ubiquitin-like protein ligase binding / RHOH GTPase cycle / polyubiquitin modification-dependent protein binding / autophagosome maturation / negative regulation of hippo signaling / endoplasmic reticulum to Golgi vesicle-mediated transport / HSF1 activation / translesion synthesis / interstrand cross-link repair / ATP metabolic process / endoplasmic reticulum unfolded protein response / proteasomal protein catabolic process / Protein methylation / Attachment and Entry / ERAD pathway / lipid droplet / proteasome complex / viral genome replication / Josephin domain DUBs / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / negative regulation of smoothened signaling pathway / macroautophagy / positive regulation of protein-containing complex assembly / Hh mutants are degraded by ERAD / establishment of protein localization / Hedgehog ligand biogenesis / Defective CFTR causes cystic fibrosis / positive regulation of non-canonical NF-kappaB signal transduction / Translesion Synthesis by POLH / ADP binding / ABC-family proteins mediated transport / autophagy / cytoplasmic stress granule / Aggrephagy / positive regulation of protein catabolic process / azurophil granule lumen / KEAP1-NFE2L2 pathway / Ovarian tumor domain proteases / positive regulation of canonical Wnt signaling pathway / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / double-strand break repair / E3 ubiquitin ligases ubiquitinate target proteins / site of double-strand break / cellular response to heat / Neddylation / ubiquitin-dependent protein catabolic process / secretory granule lumen / protein phosphatase binding / regulation of apoptotic process / ficolin-1-rich granule lumen / proteasome-mediated ubiquitin-dependent protein catabolic process / Attachment and Entry / protein ubiquitination / ciliary basal body / protein domain specific binding / DNA repair / intracellular membrane-bounded organelle / lipid binding / ubiquitin protein ligase binding / DNA damage response / Neutrophil degranulation / endoplasmic reticulum membrane / perinuclear region of cytoplasm / glutamatergic synapse / endoplasmic reticulum / protein-containing complex / ATP hydrolysis activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Yu G / Bai Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: iScience / Year: 2021 Journal: iScience / Year: 2021Title: Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation. Authors: Guimei Yu / Yunpeng Bai / Kunpeng Li / Ovini Amarasinghe / Wen Jiang / Zhong-Yin Zhang /  Abstract: VCP/p97 is an evolutionarily conserved AAA+ ATPase important for cellular homeostasis. Previous studies suggest that VCP predominantly exists as a homohexamer. Here, we performed structural and ...VCP/p97 is an evolutionarily conserved AAA+ ATPase important for cellular homeostasis. Previous studies suggest that VCP predominantly exists as a homohexamer. Here, we performed structural and biochemical characterization of VCP dodecamer, an understudied state of VCP. The structure revealed an apo nucleotide status that has rarely been captured, a tail-to-tail assembly of two hexamers, and the up-elevated N-terminal domains akin to that seen in the ATP-bound hexamer. Further analyses elucidated a nucleotide status-dependent dodecamerization mechanism, where nucleotide dissociation from the D2 AAA domains induces and promotes VCP dodecamerization. In contrast, nucleotide-free D1 AAA domains are associated with the up-rotation of N-terminal domains, which may prime D1 for ATP binding. These results therefore reveal new nucleotide status-dictated intra- and interhexamer conformational changes and suggest that modulation of D2 domain nucleotide occupancy may serve as a mechanism in controlling VCP oligomeric states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22678.map.gz emd_22678.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22678-v30.xml emd-22678-v30.xml emd-22678.xml emd-22678.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22678_fsc.xml emd_22678_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22678.png emd_22678.png | 96.2 KB | ||
| Masks |  emd_22678_msk_1.map emd_22678_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22678.cif.gz emd-22678.cif.gz | 6.6 KB | ||
| Others |  emd_22678_half_map_1.map.gz emd_22678_half_map_1.map.gz emd_22678_half_map_2.map.gz emd_22678_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22678 http://ftp.pdbj.org/pub/emdb/structures/EMD-22678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22678 | HTTPS FTP |
-Validation report
| Summary document |  emd_22678_validation.pdf.gz emd_22678_validation.pdf.gz | 943.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22678_full_validation.pdf.gz emd_22678_full_validation.pdf.gz | 942.7 KB | Display | |
| Data in XML |  emd_22678_validation.xml.gz emd_22678_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_22678_validation.cif.gz emd_22678_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22678 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22678 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22678 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22678 | HTTPS FTP |
-Related structure data
| Related structure data |  7k59MC  7k56C  7k57C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22678.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22678.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM density map of apo VCP hexamer generated from bacterially recombinant VCP/p97. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5525 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
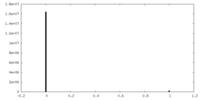
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22678_msk_1.map emd_22678_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
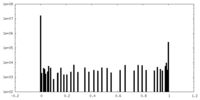
| Density Histograms |
-Half map: half map 1
| File | emd_22678_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_22678_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VCP/p97
| Entire | Name: VCP/p97 |
|---|---|
| Components |
|
-Supramolecule #1: VCP/p97
| Supramolecule | Name: VCP/p97 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: VCP/p97 was overexpressed and purified from E.coli BL21(DE3). Purified protein was treated with apyrase to remove prebound ADP and generate apo VCP/p97. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: Transitional endoplasmic reticulum ATPase
| Macromolecule | Name: Transitional endoplasmic reticulum ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: vesicle-fusing ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.464828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASGADSKGD DLSTAILKQK NRPNRLIVDE AINEDNSVVS LSQPKMDELQ LFRGDTVLLK GKKRREAVCI VLSDDTCSDE KIRMNRVVR NNLRVRLGDV ISIQPCPDVK YGKRIHVLPI DDTVEGITGN LFEVYLKPYF LEAYRPIRKG DIFLVRGGMR A VEFKVVET ...String: MASGADSKGD DLSTAILKQK NRPNRLIVDE AINEDNSVVS LSQPKMDELQ LFRGDTVLLK GKKRREAVCI VLSDDTCSDE KIRMNRVVR NNLRVRLGDV ISIQPCPDVK YGKRIHVLPI DDTVEGITGN LFEVYLKPYF LEAYRPIRKG DIFLVRGGMR A VEFKVVET DPSPYCIVAP DTVIHCEGEP IKREDEEESL NEVGYDDIGG CRKQLAQIKE MVELPLRHPA LFKAIGVKPP RG ILLYGPP GTGKTLIARA VANETGAFFF LINGPEIMSK LAGESESNLR KAFEEAEKNA PAIIFIDELD AIAPKREKTH GEV ERRIVS QLLTLMDGLK QRAHVIVMAA TNRPNSIDPA LRRFGRFDRE VDIGIPDATG RLEILQIHTK NMKLADDVDL EQVA NETHG HVGADLAALC SEAALQAIRK KMDLIDLEDE TIDAEVMNSL AVTMDDFRWA LSQSNPSALR ETVVEVPQVT WEDIG GLED VKRELQELVQ YPVEHPDKFL KFGMTPSKGV LFYGPPGCGK TLLAKAIANE CQANFISIKG PELLTMWFGE SEANVR EIF DKARQAAPCV LFFDELDSIA KARGGNIGDG GGAADRVINQ ILTEMDGMST KKNVFIIGAT NRPDIIDPAI LRPGRLD QL IYIPLPDEKS RVAILKANLR KSPVAKDVDL EFLAKMTNGF SGADLTEICQ RACKLAIRES IESEIRRERE RQTNPSAM E VEEDDPVPEI RRDHFEEAMR FARRSVSDND IRKYEMFAQT LQQSRGFGDF RFPSGNQGGA GPSQGSGGGT GGSVYTEDN DDDLYG UniProtKB: Transitional endoplasmic reticulum ATPase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 20mM Hepes pH7.4, 150mM NaCl, 1mM TCEP | ||||||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: LACEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: GRAPHENE OXIDE / Support film - #1 - topology: CONTINUOUS | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 293 K / Instrument: GATAN CRYOPLUNGE 3 Details: 3.5ul sample was applied to a lacy carbon grid coated with graphene oxide. 7 seconds blot with filter paper was performed using Gatan Cp3.. | ||||||||||||
| Details | VCP/p97 was overexpressed and purified with E.coli BL21(DE3). Purified proteins were treated with apyrase to remove prebound ADP and generate apo VCP/p97. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 807 / Average exposure time: 8.0 sec. / Average electron dose: 40.0 e/Å2 Details: A frame rate of 5 frames per second, a dose rate of 7.6 eps and a total exposure of 8 seconds were used. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)