+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22518 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SpCas9 delta4CE Ternary Complex | |||||||||
 Map data Map data | SpCas9 delta4CE Ternary Complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / DNA-binding / engineered / RNA BINDING PROTEIN | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) / Streptococcus pyogenes (bacteria) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Sham A / Laughlin TG / Savage DF | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Comprehensive deletion landscape of CRISPR-Cas9 identifies minimal RNA-guided DNA-binding modules. Authors: Arik Shams / Sean A Higgins / Christof Fellmann / Thomas G Laughlin / Benjamin L Oakes / Rachel Lew / Shin Kim / Maria Lukarska / Madeline Arnold / Brett T Staahl / Jennifer A Doudna / David F Savage /  Abstract: Proteins evolve through the modular rearrangement of elements known as domains. Extant, multidomain proteins are hypothesized to be the result of domain accretion, but there has been limited ...Proteins evolve through the modular rearrangement of elements known as domains. Extant, multidomain proteins are hypothesized to be the result of domain accretion, but there has been limited experimental validation of this idea. Here, we introduce a technique for genetic minimization by iterative size-exclusion and recombination (MISER) for comprehensively making all possible deletions of a protein. Using MISER, we generate a deletion landscape for the CRISPR protein Cas9. We find that the catalytically-dead Streptococcus pyogenes Cas9 can tolerate large single deletions in the REC2, REC3, HNH, and RuvC domains, while still functioning in vitro and in vivo, and that these deletions can be stacked together to engineer minimal, DNA-binding effector proteins. In total, our results demonstrate that extant proteins retain significant modularity from the accretion process and, as genetic size is a major limitation for viral delivery systems, establish a general technique to improve genome editing and gene therapy-based therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22518.map.gz emd_22518.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22518-v30.xml emd-22518-v30.xml emd-22518.xml emd-22518.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22518_fsc.xml emd_22518_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22518.png emd_22518.png | 93.9 KB | ||
| Masks |  emd_22518_msk_1.map emd_22518_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_22518_additional_1.map.gz emd_22518_additional_1.map.gz emd_22518_additional_2.map.gz emd_22518_additional_2.map.gz emd_22518_additional_3.map.gz emd_22518_additional_3.map.gz emd_22518_half_map_1.map.gz emd_22518_half_map_1.map.gz emd_22518_half_map_2.map.gz emd_22518_half_map_2.map.gz | 4.3 MB 6 MB 5.3 MB 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22518 http://ftp.pdbj.org/pub/emdb/structures/EMD-22518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22518 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10508 (Title: Cryo-EM structure of the SpCas9 delta4CE Ternary Complex EMPIAR-10508 (Title: Cryo-EM structure of the SpCas9 delta4CE Ternary ComplexData size: 1.5 TB Data #1: Unaligned multi-frame, non-gain-corrected frames in LZW-TIFF of SpCas9 delta4CE Ternary Complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22518.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22518.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SpCas9 delta4CE Ternary Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22518_msk_1.map emd_22518_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
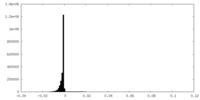
| Density Histograms |
-Additional map: #3
| File | emd_22518_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
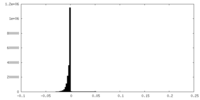
| Density Histograms |
-Additional map: #1
| File | emd_22518_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
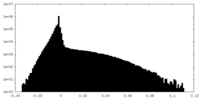
| Density Histograms |
-Additional map: #2
| File | emd_22518_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_22518_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_22518_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SpCas9 delta4CE Ternary Complex
| Entire | Name: SpCas9 delta4CE Ternary Complex |
|---|---|
| Components |
|
-Supramolecule #1: SpCas9 delta4CE Ternary Complex
| Supramolecule | Name: SpCas9 delta4CE Ternary Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 176.65 KDa |
-Macromolecule #1: SpCas9 delta4CE
| Macromolecule | Name: SpCas9 delta4CE / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAT PKKKRKVGGS PKKKRKVGGS PKKKRKVGGS PKKKRKVGIH GVPAATMDKK YSIGLAIGTN SVGWAVITDE YKVPSKKFKV LGNTDRHSIK KNLIGALLFD SGETAEATRL KRTARRRYTR RKNRICYLQE IFSNEMAKVD DSFFHRLEES ...String: MKSSHHHHHH ENLYFQSNAT PKKKRKVGGS PKKKRKVGGS PKKKRKVGGS PKKKRKVGIH GVPAATMDKK YSIGLAIGTN SVGWAVITDE YKVPSKKFKV LGNTDRHSIK KNLIGALLFD SGETAEATRL KRTARRRYTR RKNRICYLQE IFSNEMAKVD DSFFHRLEES FLVEEDKKHE RHPIFGNIVD EVAYHEKYPT IYHLRKKLVD STDKADLRLI YLALAHMIKF RGHFLIEGDL NPDNSTSDAI LLSDILRVNT EITKAPLSAS MIKRYDEHHQ DLTLLKALVR QQLPEKYKEI FFDQSKNGYA GYIDGGASQE EFYKFIKPIL EKMDGTEELL VKLNREDLLR KQRTFDNGSI PHQIHLGELH AILRRQEDFY PFLKDNREKI EKILTFRIPY YVGPLARGNS RFAWMTRKSE ETITPWNFEE VVDKGASAQS FIERMTNFDK NLTSQKAQVS GQGDSLHEHI ANLAGSPAIK KGILQTVKVV DELVKVMGRH KPENIVIEMA RENQTTQKGQ KNSRERMKRI EEGIKELASD NLTKAERGGL SELDKAGFIK RQLVETRQIT KHVAQILDSR MNTKYDENDK LIREVKVITL KSKLVSDFRK DFQFYKVREI NNYHHAHDAY LNAVVGTALI KKYPKLESEF VASTVRKVLS MPQVNIVKKT EVQTGGFSKE SILPKRNSDK LIARKKDWDP KKYGGFDSPT VAYSVLVVAK VEKGKSKKLK SVKELLGITI MERSSFEKNP IDFLEAKGYK EVKKDLIIKL PKYSLFELEN GRKRMLASAG ELQKGNELAL PSKYVNFLYL ASHYEKLKGS PEDNEQKQLF VEQHKHYLDE IIEQISEFSK RVILADANLD KVLSAYNKHR DKPIREQAEN IIHLFTLTNL GAPAAFKYFD TTIDRKRYTS TKEVLDATLI HQSITGLYET RIDLSQLGGD GSPKKKRKVE DPKKKRKVDT QSPKGS |
-Macromolecule #2: sgRNA
| Macromolecule | Name: sgRNA / type: rna / ID: 2 |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: UCAGCCACAG CUGGGCCUGG GUUUUAGAGC UAGAAAUAGC AAGUUAAAAU AAGGCUAGUC CGUUAUCAAC UUGAAAAAGU GGCACCGAGU CGGUGCUUUU |
-Macromolecule #3: Target-strand
| Macromolecule | Name: Target-strand / type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: CTCACCTTCC TACGGTAGTC GGTGTCGACC CGGACCACCC GACAGTTTTA ACTCG |
-Macromolecule #4: Non-target strand
| Macromolecule | Name: Non-target strand / type: dna / ID: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: GAGTGGAAGG ATGCCATCAG CCACAGCTGG GCCTGGTGGG CTGTCAAAAT TGAGC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0053 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV / Details: 10 s incubation, blot 5 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3400 / Average electron dose: 60.0 e/Å2 Details: collected using image-shift for a regular 3x3 array of holes per stage movement |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.8000000000000003 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 395 / Target criteria: CC |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)