[English] 日本語
 Yorodumi
Yorodumi- EMDB-22468: 70S ribosome stalled on long mRNA with ArfB C-terminus bound in t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22468 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 70S ribosome stalled on long mRNA with ArfB C-terminus bound in the mRNA tunnel | |||||||||
 Map data Map data | Refinement map for fit 9-I B factor softened by 50 angstroms squared. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Carbone CE / Korostelev AA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: ArfB can displace mRNA to rescue stalled ribosomes. Authors: Christine E Carbone / Gabriel Demo / Rohini Madireddy / Egor Svidritskiy / Andrei A Korostelev /   Abstract: Ribosomes stalled during translation must be rescued to replenish the pool of translation-competent ribosomal subunits. Bacterial alternative rescue factor B (ArfB) releases nascent peptides from ...Ribosomes stalled during translation must be rescued to replenish the pool of translation-competent ribosomal subunits. Bacterial alternative rescue factor B (ArfB) releases nascent peptides from ribosomes stalled on mRNAs truncated at the A site, allowing ribosome recycling. Prior structural work revealed that ArfB recognizes such ribosomes by inserting its C-terminal α-helix into the vacant mRNA tunnel. In this work, we report that ArfB can efficiently recognize a wider range of mRNA substrates, including longer mRNAs that extend beyond the A-site codon. Single-particle cryo-EM unveils that ArfB employs two modes of function depending on the mRNA length. ArfB acts as a monomer to accommodate a shorter mRNA in the ribosomal A site. By contrast, longer mRNAs are displaced from the mRNA tunnel by more than 20 Å and are stabilized in the intersubunit space by dimeric ArfB. Uncovering distinct modes of ArfB function resolves conflicting biochemical and structural studies, and may lead to re-examination of other ribosome rescue pathways, whose functions depend on mRNA lengths. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22468.map.gz emd_22468.map.gz | 164.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22468-v30.xml emd-22468-v30.xml emd-22468.xml emd-22468.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22468.png emd_22468.png | 98.2 KB | ||
| Others |  emd_22468_additional_1.map.gz emd_22468_additional_1.map.gz emd_22468_half_map_1.map.gz emd_22468_half_map_1.map.gz emd_22468_half_map_2.map.gz emd_22468_half_map_2.map.gz | 164.8 MB 83.8 MB 83.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22468 http://ftp.pdbj.org/pub/emdb/structures/EMD-22468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22468 | HTTPS FTP |
-Related structure data
| Related structure data |  7jssC  7jswC  7jszC  7jt1C  7jt2C  7jt3C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22468.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22468.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map for fit 9-I B factor softened by 50 angstroms squared. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.042 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Original 3.4 angstrom map with no B factor applied.
| File | emd_22468_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Original 3.4 angstrom map with no B factor applied. | ||||||||||||
| Projections & Slices |
| ||||||||||||
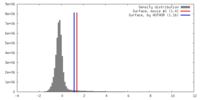
| Density Histograms |
-Half map: half map 1
| File | emd_22468_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
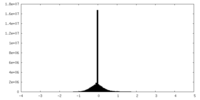
| Density Histograms |
-Half map: half map 2
| File | emd_22468_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 70S ribosome stalled on long mRNA with ArfB C-terminus bound in t...
| Entire | Name: 70S ribosome stalled on long mRNA with ArfB C-terminus bound in the mRNA tunnel |
|---|---|
| Components |
|
-Supramolecule #1: 70S ribosome stalled on long mRNA with ArfB C-terminus bound in t...
| Supramolecule | Name: 70S ribosome stalled on long mRNA with ArfB C-terminus bound in the mRNA tunnel type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 49.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Ab initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 9841 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)