[English] 日本語
 Yorodumi
Yorodumi- EMDB-22381: Structure of the activated Roq1 resistosome directly recognizing ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22381 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | |||||||||
 Map data Map data | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Resistosome / Plant Immunity / Effector / LRR / TIR / NB-ARC / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / NAD+ nucleosidase activity, cyclic ADP-ribose generating / Hydrolases; Glycosylases; Hydrolysing N-glycosyl compounds / defense response / ADP binding / signal transduction Similarity search - Function | |||||||||
| Biological species |   Xanthomonas euvesicatoria (bacteria) Xanthomonas euvesicatoria (bacteria) | |||||||||
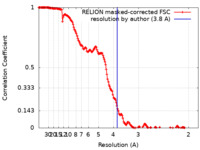
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Martin R / Qi T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Authors: Raoul Martin / Tiancong Qi / Haibo Zhang / Furong Liu / Miles King / Claire Toth / Eva Nogales / Brian J Staskawicz /   Abstract: Plants and animals detect pathogen infection using intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) that directly or indirectly recognize pathogen effectors and activate an ...Plants and animals detect pathogen infection using intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) that directly or indirectly recognize pathogen effectors and activate an immune response. How effector sensing triggers NLR activation remains poorly understood. Here we describe the 3.8-angstrom-resolution cryo-electron microscopy structure of the activated ROQ1 (recognition of XopQ 1), an NLR native to with a Toll-like interleukin-1 receptor (TIR) domain bound to the effector XopQ ( outer protein Q). ROQ1 directly binds to both the predicted active site and surface residues of XopQ while forming a tetrameric resistosome that brings together the TIR domains for downstream immune signaling. Our results suggest a mechanism for the direct recognition of effectors by NLRs leading to the oligomerization-dependent activation of a plant resistosome and signaling by the TIR domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22381.map.gz emd_22381.map.gz | 395.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22381-v30.xml emd-22381-v30.xml emd-22381.xml emd-22381.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22381_fsc.xml emd_22381_fsc.xml | 17.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_22381.png emd_22381.png | 105.5 KB | ||
| Filedesc metadata |  emd-22381.cif.gz emd-22381.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22381 http://ftp.pdbj.org/pub/emdb/structures/EMD-22381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22381 | HTTPS FTP |
-Validation report
| Summary document |  emd_22381_validation.pdf.gz emd_22381_validation.pdf.gz | 703.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22381_full_validation.pdf.gz emd_22381_full_validation.pdf.gz | 702.8 KB | Display | |
| Data in XML |  emd_22381_validation.xml.gz emd_22381_validation.xml.gz | 15.1 KB | Display | |
| Data in CIF |  emd_22381_validation.cif.gz emd_22381_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22381 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22381 | HTTPS FTP |
-Related structure data
| Related structure data |  7jlvMC  7jluC  7jlxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22381.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22381.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9386 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Roq1
| Entire | Name: Roq1 |
|---|---|
| Components |
|
-Supramolecule #1: Roq1
| Supramolecule | Name: Roq1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.819 KDa |
-Supramolecule #2: XopQ
| Supramolecule | Name: XopQ / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Xanthomonas euvesicatoria (bacteria) Xanthomonas euvesicatoria (bacteria) |
-Macromolecule #1: Disease resistance protein Roq1
| Macromolecule | Name: Disease resistance protein Roq1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150.648391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLTSSSHHGR SYDVFLSFRG EDTRKTFVGH LFNALIEKGI HTFMDDKELK RGKSISSELM KAIGESRFAV VVFSKNYASS TWCLEELVK ILEIHEKFEL IVVPVFYDVD PSTVRKQNGE YAVCFTKFEA NLVDDRDKVL RWREALTKVA NISGHDLRNT Y NGDESKCI ...String: MLTSSSHHGR SYDVFLSFRG EDTRKTFVGH LFNALIEKGI HTFMDDKELK RGKSISSELM KAIGESRFAV VVFSKNYASS TWCLEELVK ILEIHEKFEL IVVPVFYDVD PSTVRKQNGE YAVCFTKFEA NLVDDRDKVL RWREALTKVA NISGHDLRNT Y NGDESKCI QQILKDIFDK FCFSISITNR DLVGIESQIK KLSSLLRMDL KGVRLVGIWG MGGVGKTTAA RALFNRYYQN FE SACFLED VKEYLQHHTL LYLQKTLLSK LLKVEFVDCT DTEEMCVILK RRLCSKKVLV VLDDVNHNDQ LDKLVGAEDW FGS GSRIVI TTRDMKLLKN HDVHETYEIK VLEKDEAIEL FNLHAFKRSS PEKEFKELLN LVVDYTGGLP LALKVLGSLL YKED LDVWI STIDRLKDNP EGEIMATLKI SFDGLRDYEK SIFLDIACFF RGYNQRDMTA LFHASGFHPV LGVKTLVEKS LIFIL EDKI QMHDLMQEMG RQIAVQESPM RRIYRPEDVK DACIGDMRKE AIEGLLLTEP EQFEEGELEY MYSAEALKKT RRLRIL VKE YYNRGFDEPV AYLPNSLLWL EWRNYSSNSF PSNFEPSKLV YLTMKGSSII ELWNGAKRLA FLTTLDLSYC HKLIQTP DF RMITNLERLI LSSCDALVEV HPSVGFLKNL ILLNMDHCIS LERLPAIIQS ECLEVLDLNY CFNLKMFPEV ERNMTHLK K LDLTSTGIRE LPASIEHLSS LENLQMHSCN QLVSLPSSIW RFRNLKISEC EKLGSLPEIH GNSNCTRELI LKLVSIKEL PTSIGNLTSL NFLEICNCKT ISSLSSSIWG LTSLTTLKLL DCRKLKNLPG IPNAINHLSG HGLQLLLTLE QPTIYERLDL LRIIDMSWC SCISSLPHNI WMLKFLRILC ISYCSRLEYL PENLGHLEHL EELLADGTGI LRLPSSVARL NKLEVLSFRK K FAIGPKVQ YSSSMLNLPD DVFGSLGSLG SVVKLNLSGN GFCNLPETMN QLFCLEYLDI TFCQRLEALP ELPPSIKELY VD EHLALRI MEDLVIKCKE LNLIAVTKIE YQNFYRWLDS IWSDVSELLE NSQKQQLDDM LQLIPFSYLS TAKREEVLKI VIH GTRIPE WFRWQDRSAT TMSVNLPEYW YTENFLGFAI CCSCCFYHSA RSYDVEFEGS MHHYNYDSSY WKEYEEPSYD FYER DSIEI TAKLTPRHKG MRTEELKKVC SFSMNVLRRA TAVPNMCFAF FPFNSLCHIS NLQANNPNDY GIFETCLSPG DIRHR GKQW GFNLVYKDET GGSVTHEMLI NR UniProtKB: Disease resistance protein Roq1 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 10 sec blot. Blot Force 10. 90 min incubation.. | ||||||||||||||
| Details | Sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 11134 / Average electron dose: 50.0 e/Å2 Details: Images were collected as dose-fractionated movie frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 80879 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







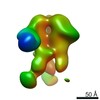

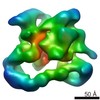



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)