+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22378 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Herpes Simplex Virus Type 1 Procapsid with Portal Vertex | |||||||||
 Map data Map data | Herpes Simplex Virus Type 1 Procapsid with Portal Vertex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) | |||||||||
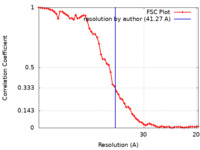
| Method | subtomogram averaging / cryo EM / Resolution: 41.27 Å | |||||||||
 Authors Authors | Buch MHC / Heymann JB / Newcomb WW / Winkler DC / Steven AC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2021 Journal: mBio / Year: 2021Title: Cryo-Electron Tomography of the Herpesvirus Procapsid Reveals Interactions of the Portal with the Scaffold and a Shift on Maturation. Authors: Michael H C Buch / William W Newcomb / Dennis C Winkler / Alasdair C Steven / J Bernard Heymann /  Abstract: Herpes simplex virus 1 (HSV-1) requires seven proteins to package its genome through a vertex in its capsid, one of which is the portal protein, pUL6. The portal protein is also thought to facilitate ...Herpes simplex virus 1 (HSV-1) requires seven proteins to package its genome through a vertex in its capsid, one of which is the portal protein, pUL6. The portal protein is also thought to facilitate assembly of the procapsid. While the portal has been visualized in mature capsids, we aimed to elucidate its role in the assembly and maturation of procapsids using cryo-electron tomography (cryoET). We identified the portal vertex in individual procapsids, calculated a subtomogram average, and compared that with the portal vertex in empty mature capsids (A-capsids). The resulting maps show the portal on the interior surface with its narrower end facing outwards, while maintaining close contact with the capsid shell. In the procapsid, the portal is embedded in the underlying scaffold, suggesting that assembly involves a portal-scaffold complex. During maturation, the capsid shell angularizes with a corresponding outward movement of the vertices. We found that in A-capsids, the portal translocates outward further than the adjacent capsomers and strengthens its contacts with the capsid shell. Our methodology also allowed us to determine the number of portal vertices in each capsid, with most having one per capsid, but some none or two, and rarely three. The predominance of a single portal per capsid supports facilitation of the assembly of the procapsid. Herpes simplex virus 1 (HSV-1) infects a majority of humans, causing mostly mild disease but in some cases progressing toward life-threatening encephalitis. Understanding the life cycle of the virus is important to devise countermeasures. Production of the virion starts with the assembly of an icosahedral procapsid, which includes DNA packaging proteins at a vertex, one of which is the dodecameric portal protein. The procapsid then undergoes maturation and DNA packaging through the portal, driven by a terminase complex. We used cryo-electron tomography to visualize the portal in procapsids and compare them to mature empty capsids. We found the portal located inside one vertex interacting with the scaffold protein in the procapsid. On maturation, the scaffold is cleaved and dissociates, the capsid angularizes, and the portal moves outward, interacting closely with the capsid shell. These transformations may provide a basis for the development of drugs to prevent HSV-1 infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22378.map.gz emd_22378.map.gz | 317.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22378-v30.xml emd-22378-v30.xml emd-22378.xml emd-22378.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22378_fsc.xml emd_22378_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22378.png emd_22378.png | 113.3 KB | ||
| Masks |  emd_22378_msk_1.map emd_22378_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Others |  emd_22378_half_map_1.map.gz emd_22378_half_map_1.map.gz emd_22378_half_map_2.map.gz emd_22378_half_map_2.map.gz | 324 MB 323.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22378 http://ftp.pdbj.org/pub/emdb/structures/EMD-22378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22378 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22378.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22378.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Herpes Simplex Virus Type 1 Procapsid with Portal Vertex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.6055 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
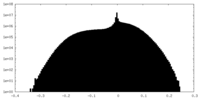
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22378_msk_1.map emd_22378_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
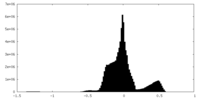
| Density Histograms |
-Half map: Herpes Simplex Virus Type 1 Procapsid with Portal Vertex
| File | emd_22378_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Herpes Simplex Virus Type 1 Procapsid with Portal Vertex | ||||||||||||
| Projections & Slices |
| ||||||||||||
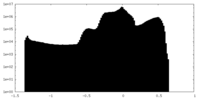
| Density Histograms |
-Half map: Herpes Simplex Virus Type 1 Procapsid with Portal Vertex
| File | emd_22378_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Herpes Simplex Virus Type 1 Procapsid with Portal Vertex | ||||||||||||
| Projections & Slices |
| ||||||||||||
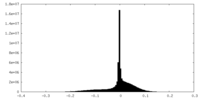
| Density Histograms |
- Sample components
Sample components
-Entire : Human alphaherpesvirus 1
| Entire | Name:   Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) |
|---|---|
| Components |
|
-Supramolecule #1: Human alphaherpesvirus 1
| Supramolecule | Name: Human alphaherpesvirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Procapsids were obtained from infected Vero cells. Purification steps were carried out at room temperature. Harvested cells were resuspended in PBS buffer and lysed by addition of TX-100. ...Details: Procapsids were obtained from infected Vero cells. Purification steps were carried out at room temperature. Harvested cells were resuspended in PBS buffer and lysed by addition of TX-100. Procapsids were purified by multiple filtration steps using Eppendorf spin filters. Turquoise pellets containing procapsids were resuspended in PBS, sonicated in a bath sonicator, and concentrated to the desired volume using a 300 kDa MWCO filter. NCBI-ID: 10298 / Sci species name: Human alphaherpesvirus 1 / Sci species strain: KOS / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Host system | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Virus shell | Shell ID: 1 / Name: Procapsid / Diameter: 1250.0 Å / T number (triangulation number): 16 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: PBS |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER Details: Grids were plasma-cleaned in a Model 1020 plasma cleaner (Fischione, Export, PA). |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: LEICA EM GP Details: Sample was mixed with Aurion Anionic 10 nm BSA gold tracer (EMS) at a 1:1 ratio, and 3 uL were applied to Quantifoil copper grids covered with 300-mesh holey carbon (EMS). Excess liquid was ...Details: Sample was mixed with Aurion Anionic 10 nm BSA gold tracer (EMS) at a 1:1 ratio, and 3 uL were applied to Quantifoil copper grids covered with 300-mesh holey carbon (EMS). Excess liquid was blotted off for 2.5 s, and grids were flash-frozen in liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-2 / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 3.1 e/Å2 Details: Tilts were imaged in movie mode with 2 movies at 0.5 seconds each. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)