+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5384 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of PBCV-1 (five-fold averaged map) | |||||||||
 Map data Map data | Five-fold averaged map of PBCV-1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cell entry / minor protein / double jelly-roll | |||||||||
| Biological species |   Paramecium bursaria Chlorella virus 1 Paramecium bursaria Chlorella virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.8 Å | |||||||||
 Authors Authors | Zhang XZ / Xiang Y / Dunigan DD / Klose T / Chipman PR / Etten JLV / Rossmann MG | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Authors: Xinzheng Zhang / Ye Xiang / David D Dunigan / Thomas Klose / Paul R Chipman / James L Van Etten / Michael G Rossmann /  Abstract: A cryoelectron microscopy 8.5 Å resolution map of the 1,900 Å diameter, icosahedral, internally enveloped Paramecium bursaria chlorella virus was used to interpret structures of the virus at ...A cryoelectron microscopy 8.5 Å resolution map of the 1,900 Å diameter, icosahedral, internally enveloped Paramecium bursaria chlorella virus was used to interpret structures of the virus at initial stages of cell infection. A fivefold averaged map demonstrated that two minor capsid proteins involved in stabilizing the capsid are missing in the vicinity of the unique vertex. Reconstruction of the virus in the presence of host chlorella cell walls established that the spike at the unique vertex initiates binding to the cell wall, which results in the enveloped nucleocapsid moving closer to the cell. This process is concurrent with the release of the internal viral membrane that was linked to the capsid by many copies of a viral membrane protein in the mature infectous virus. Simultaneously, part of the trisymmetrons around the unique vertex disassemble, probably in part because two minor capsid proteins are absent, causing Paramecium bursaria chlorella virus and the cellular contents to merge, possibly as a result of enzyme(s) within the spike assembly. This may be one of only a few recordings of successive stages of a virus while infecting a eukaryotic host in pseudoatomic detail in three dimensions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5384.map.gz emd_5384.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5384-v30.xml emd-5384-v30.xml emd-5384.xml emd-5384.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5384_1.jpg emd_5384_1.jpg | 98.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5384 http://ftp.pdbj.org/pub/emdb/structures/EMD-5384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5384 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5384.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5384.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
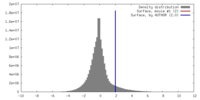
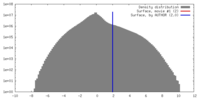
| Annotation | Five-fold averaged map of PBCV-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PBCV-1
| Entire | Name: PBCV-1 |
|---|---|
| Components |
|
-Supramolecule #1000: PBCV-1
| Supramolecule | Name: PBCV-1 / type: sample / ID: 1000 / Number unique components: 8 |
|---|
-Supramolecule #1: Paramecium bursaria Chlorella virus 1
| Supramolecule | Name: Paramecium bursaria Chlorella virus 1 / type: virus / ID: 1 / Name.synonym: PBCV-1 / NCBI-ID: 10506 / Sci species name: Paramecium bursaria Chlorella virus 1 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: PBCV-1 |
|---|---|
| Host (natural) | Organism:  Chlorella variabilis (plant) / synonym: ALGAE Chlorella variabilis (plant) / synonym: ALGAE |
| Virus shell | Shell ID: 1 / Name: VP54 / Diameter: 1850 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: BBM |
| Grid | Details: 200 mesh grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 174 K / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Temperature | Average: 175 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 250,000 times magnification |
| Image recording | Category: CCD / Film or detector model: KODAK SO-163 FILM / Digitization - Sampling interval: 6.75 µm / Number real images: 800 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 33065 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder: Single tilt holder / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each micrograph |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 17016 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)