+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22339 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM map of Aureococcus anophagefferens Virus (AaV) | |||||||||
 Map data Map data | Map of the capsid of Aureococcus anophagefferens Virus | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Aureococcus anophagefferens virus Aureococcus anophagefferens virus | |||||||||
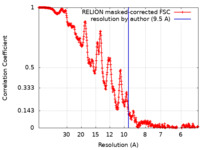
| Method | single particle reconstruction / cryo EM / Resolution: 9.5 Å | |||||||||
 Authors Authors | Gann ER / Xian Y / Abraham PE / Hettich RL / Reynolds TB / Xiao C / Wilhelm SW | |||||||||
 Citation Citation |  Journal: Front Microbiol / Year: 2020 Journal: Front Microbiol / Year: 2020Title: Structural and Proteomic Studies of the Virus Demonstrate a Global Distribution of Virus-Encoded Carbohydrate Processing. Authors: Eric R Gann / Yuejiao Xian / Paul E Abraham / Robert L Hettich / Todd B Reynolds / Chuan Xiao / Steven W Wilhelm /  Abstract: Viruses modulate the function(s) of environmentally relevant microbial populations, yet considerations of the metabolic capabilities of individual virus particles themselves are rare. We used shotgun ...Viruses modulate the function(s) of environmentally relevant microbial populations, yet considerations of the metabolic capabilities of individual virus particles themselves are rare. We used shotgun proteomics to quantitatively identify 43 virus-encoded proteins packaged within purified Virus (AaV) particles, normalizing data to the per-virion level using a 9.5-Å-resolution molecular reconstruction of the 1900-Å (AaV) particle that we generated with cryogenic electron microscopy. This packaged proteome was used to determine similarities and differences between members of different giant virus families. We noted that proteins involved in sugar degradation and binding (e.g., carbohydrate lyases) were unique to AaV among characterized giant viruses. To determine the extent to which this virally encoded metabolic capability was ecologically relevant, we examined the TARA Oceans dataset and identified genes and transcripts of viral origin. Our analyses demonstrated that putative giant virus carbohydrate lyases represented up to 17% of the marine pool for this function. In total, our observations suggest that the AaV particle has potential prepackaged metabolic capabilities and that these may be found in other giant viruses that are widespread and abundant in global oceans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22339.map.gz emd_22339.map.gz | 139.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22339-v30.xml emd-22339-v30.xml emd-22339.xml emd-22339.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22339_fsc.xml emd_22339_fsc.xml | 36.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22339.png emd_22339.png | 219 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22339 http://ftp.pdbj.org/pub/emdb/structures/EMD-22339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22339 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22339.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22339.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the capsid of Aureococcus anophagefferens Virus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.68 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Aureococcus anophagefferens virus
| Entire | Name:  Aureococcus anophagefferens virus Aureococcus anophagefferens virus |
|---|---|
| Components |
|
-Supramolecule #1: Aureococcus anophagefferens virus
| Supramolecule | Name: Aureococcus anophagefferens virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 1474867 / Sci species name: Aureococcus anophagefferens virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Aureococcus anophagefferens (eukaryote) Aureococcus anophagefferens (eukaryote) |
| Virus shell | Shell ID: 1 / Diameter: 1900.0 Å / T number (triangulation number): 169 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 50 mM Tris base, 250 mM sodium chloride, 5 mM magnesium chloride |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 6.842 µm / Calibrated defocus min: 0.493 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)