[English] 日本語
 Yorodumi
Yorodumi- EMDB-22327: Cryo-EM structure of P. falciparum VAR2CSA NF54 core in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22327 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of P. falciparum VAR2CSA NF54 core in complex with CSA at 3.36 A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Placental malaria / VAR2CSA / Duffy binding domain / CSA / PfEMP1 / SUGAR BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Ma R / Tolia NH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2021 Journal: Nat Microbiol / Year: 2021Title: Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA. Authors: Rui Ma / Tengfei Lian / Rick Huang / Jonathan P Renn / Jennifer D Petersen / Joshua Zimmerberg / Patrick E Duffy / Niraj H Tolia /  Abstract: Plasmodium falciparum VAR2CSA binds to chondroitin sulfate A (CSA) on the surface of the syncytiotrophoblast during placental malaria. This interaction facilitates placental sequestration of malaria ...Plasmodium falciparum VAR2CSA binds to chondroitin sulfate A (CSA) on the surface of the syncytiotrophoblast during placental malaria. This interaction facilitates placental sequestration of malaria parasites resulting in severe health outcomes for both the mother and her offspring. Furthermore, CSA is presented by diverse cancer cells and specific targeting of cells by VAR2CSA may become a viable approach for cancer treatment. In the present study, we determined the cryo-electron microscopy structures of the full-length ectodomain of VAR2CSA from P. falciparum strain NF54 in complex with CSA, and VAR2CSA from a second P. falciparum strain FCR3. The architecture of VAR2CSA is composed of a stable core flanked by a flexible arm. CSA traverses the core domain by binding within two channels and CSA binding does not induce major conformational changes in VAR2CSA. The CSA-binding elements are conserved across VAR2CSA variants and are flanked by polymorphic segments, suggesting immune selection outside the CSA-binding sites. This work provides paths for developing interventions against placental malaria and cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22327.map.gz emd_22327.map.gz | 92.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22327-v30.xml emd-22327-v30.xml emd-22327.xml emd-22327.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22327.png emd_22327.png | 57.6 KB | ||
| Filedesc metadata |  emd-22327.cif.gz emd-22327.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22327 http://ftp.pdbj.org/pub/emdb/structures/EMD-22327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22327 | HTTPS FTP |
-Related structure data
| Related structure data |  7jghMC  7jgdC  7jgeC  7jgfC  7jggC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22327.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22327.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : VAR2CSA NF54 core in complex with CSA
| Entire | Name: VAR2CSA NF54 core in complex with CSA |
|---|---|
| Components |
|
-Supramolecule #1: VAR2CSA NF54 core in complex with CSA
| Supramolecule | Name: VAR2CSA NF54 core in complex with CSA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Erythrocyte membrane protein 1
| Macromolecule | Name: Erythrocyte membrane protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 308.706031 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TGMDKSSIAN KIEAYLGAKS DDSKIDQSLK ADPSEVQYYG SGGDGYYLRK NICKITVNHS DSGTNDPCDR IPPPYGDNDQ WKCAIILSK VSEKPENVFV PPRRQRMCIN NLEKLNVDKI RDKHAFLADV LLTARNEGER IVQNHPDTNS SNVCNALERS F ADIADIIR ...String: TGMDKSSIAN KIEAYLGAKS DDSKIDQSLK ADPSEVQYYG SGGDGYYLRK NICKITVNHS DSGTNDPCDR IPPPYGDNDQ WKCAIILSK VSEKPENVFV PPRRQRMCIN NLEKLNVDKI RDKHAFLADV LLTARNEGER IVQNHPDTNS SNVCNALERS F ADIADIIR GTDLWKGTNS NLEQNLKQMF AKIRENDKVL QDKYPKDQNY RKLREDWWNA NRQKVWEVIT CGARSNDLLI KR GWRTSGK SNGDNKLELC RKCGHYEEKV PTKLDYVPQF LRWLTEWIED FYREKQNLID DMERHREECT SEDHKSKEGT SYC STCKDK CKKYCECVKK WKSEWENQKN KYTELYQQNK NETSQKNTSR YDDYVKDFFK KLEANYSSLE NYIKGDPYFA EYAT KLSFI LNSSDANNPS EKIQKNNDEV CNCNESGIAS VEQEQISDPS SNKTCITHSS IKANKKKVCK HVKLGVREND KDLRV CVIE HTSLSGVENC CCQDFLRILQ ENCSDNKSGS SSNGSCNNKN QEACEKNLEK VLASLTNCYK CDKCKSEQSK KNNKNW IWK KSSGKEGGLQ KEYANTIGLP PRTQSLCLVV CLDEKGKKTQ ELKNIRTNSE LLKEWIIAAF HEGKNLKPSH EKKNDDN GK KLCKALEYSF ADYGDLIKGT SIWDNEYTKD LELNLQKIFG KLFRKYIKKN NTAEQDTSYS SLDELRESWW NTNKKYIW L AMKHGAGMNS TTCCGDGSVT GSGSSCDDIP TIDLIPQYLR FLQEWVEHFC KQRQEKVKPV IENCKSCKES GGTCNGECK TECKNKCEVY KKFIEDCKGG DGTAGSSWVK RWDQIYKRYS KYIEDAKRNR KAGTKNCGPS STTNAAENKC VQSDIDSFFK HLIDIGLTT PSSYLSIVLD DNICGADKAP WTTYTTYTTT EKCNKETDKS KLQQCNTAVV VNVPSPLGNT PHGYKYACQC K IPTNEETC DDRKEYMNQW SCGSARTMKR GYKNDNYELC KYNGVDVKPT TVRSNSSKLD DKDVTFFNLF EQWNKEIQYQ IE QYMTNTK ISCNNEKNVL SRVSDEAAQP KFSDNERDRN SITHEDKNCK EKCKCYSLWI EKINDQWDKQ KDNYNKFQRK QIY DANKGS QNKKVVSLSN FLFFSCWEEY IQKYFNGDWS KIKNIGSDTF EFLIKKCGND SGDGETIFSE KLNNAEKKCK ENES TNNKM KSSETSCDCS EPIYIRGCQP KIYDGKIFPG KGGEKQWICK DTIIHGDTNG ACIPPRTQNL CVGELWDKRY GGRSN IKND TKESLKQKIK NAIQKETELL YEYHDKGTAI ISRNPMKGQK EKEEKNNDSN GLPKGFCHAV QRSFIDYKNM ILGTSV NIY EYIGKLQEDI KKIIEKGTTK QNGKTVGSGA ENVNAWWKGI EGEMWDAVRC AITKINKKQK KNGTFSIDEC GIFPPTG ND EDQSVSWFKE WSEQFCIERL QYEKNIRDAC TNNGQGDKIQ GDCKRKCEEY KKYISEKKQE WDKQKTKYEN KYVGKSAS D LLKENYPECI SANFDFIFND NIEYKTYYPY GDYSSICSCE QVKYYEYNNA EKKNNKSLCH EKGNDRTWSK KYIKKLENG RTLEGVYVPP RRQQLCLYEL FPIIIKNKND ITNAKKELLE TLQIVAEREA YYLWKQYHAH NDTTYLAHKK ACCAIRGSFY DLEDIIKGN DLVHDEYTKY IDSKLNEIFD SSNKNDIETK RARTDWWENE AIAVPNITGA NKSDPKTIRQ LVWDAMQSGV R KAIDEEKE KKKPNENFPP CMGVQHIGIA KPQFIRWLEE WTNEFCEKYT KYFEDMKSNC NLRKGADDCD DNSNIECKKA CA NYTNWLN PKRIEWNGMS NYYNKIYRKS NKESEDGKDY SMIMEPTVID YLNKRCNGEI NGNYICCSCK NIGENSTSGT VNK KLQKKE TQCEDNKGPL DLMNKVLNKM DPKYSEHKMK CTEVYLEHVE EQLKEIDNAI KDYKLYPLDR CFDDKSKMKV CDLI GDAIG CKHKTKLDEL DEWNDVDMRD PYNKYKGVLI PPRRRQLCFS RIVRGPANLR NLKEFKEEIL KGAQSEGKFL GNYYN EDKD KEKALEAMKN SFYDYEYIIK GSDMLTNIQF KDIKRKLDRL LEKETNNTEK VDDWWETNKK SIWNAMLCGY KKSGNK IID PSWCTIPTTE TPPQFLRWIK EWGTNVCIQK EEHKEYVKSK CSNVTNLGAQ ESESKNCTSE IKKYQEWSRK RSIQWEA IS EGYKKYKGMD EFKNTFKNIK EPDANEPNAN EYLKKHCSKC PCGFNDMQEI TKYTNIGNEA FKQIKEQVDI PAELEDVI Y RLKHHEYDKG NDYICNKYKN INVNMKKNND DTWTDLVKNS SDINKGVLLP PRRKNLFLKI DESDICKYKR DPKLFKDFI YSSAISEVER LKKVYGEAKT KVVHAMKYSF ADIGSIIKGD DMMENNSSDK IGKILGDGVG QNEKRKKWWD MNKYHIWESM LCGYKHAYG NISENDRKML DIPNNDDEHQ FLRWFQEWTE NFCTKRNELY ENMVTACNSA KCNTSNGSVD KKECTEACKN Y SNFILIKK KEYQSLNSQY DMNYKETKAE KKESPEYFKD KCNGECSCLS EYFKDETRWK NPYETLDDTE VKNNCMCKPP PP ASNNGTK HHHHHH UniProtKB: Erythrocyte membrane protein 1 |
-Macromolecule #3: 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: ASG |
|---|---|
| Molecular weight | Theoretical: 301.271 Da |
| Chemical component information | 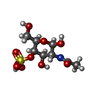 ChemComp-ASG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















