+Search query
-Structure paper
| Title | Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA. |
|---|---|
| Journal, issue, pages | Nat Microbiol, Vol. 6, Issue 3, Page 380-391, Year 2021 |
| Publish date | Jan 15, 2021 |
 Authors Authors | Rui Ma / Tengfei Lian / Rick Huang / Jonathan P Renn / Jennifer D Petersen / Joshua Zimmerberg / Patrick E Duffy / Niraj H Tolia /  |
| PubMed Abstract | Plasmodium falciparum VAR2CSA binds to chondroitin sulfate A (CSA) on the surface of the syncytiotrophoblast during placental malaria. This interaction facilitates placental sequestration of malaria ...Plasmodium falciparum VAR2CSA binds to chondroitin sulfate A (CSA) on the surface of the syncytiotrophoblast during placental malaria. This interaction facilitates placental sequestration of malaria parasites resulting in severe health outcomes for both the mother and her offspring. Furthermore, CSA is presented by diverse cancer cells and specific targeting of cells by VAR2CSA may become a viable approach for cancer treatment. In the present study, we determined the cryo-electron microscopy structures of the full-length ectodomain of VAR2CSA from P. falciparum strain NF54 in complex with CSA, and VAR2CSA from a second P. falciparum strain FCR3. The architecture of VAR2CSA is composed of a stable core flanked by a flexible arm. CSA traverses the core domain by binding within two channels and CSA binding does not induce major conformational changes in VAR2CSA. The CSA-binding elements are conserved across VAR2CSA variants and are flanked by polymorphic segments, suggesting immune selection outside the CSA-binding sites. This work provides paths for developing interventions against placental malaria and cancer. |
 External links External links |  Nat Microbiol / Nat Microbiol /  PubMed:33452495 / PubMed:33452495 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.36 - 4.88 Å |
| Structure data | EMDB-22323, PDB-7jgd: EMDB-22324, PDB-7jge: EMDB-22325, PDB-7jgf: EMDB-22326, PDB-7jgg: EMDB-22327, PDB-7jgh: |
| Chemicals | 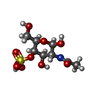 ChemComp-ASG: |
| Source |
|
 Keywords Keywords | SUGAR BINDING PROTEIN / Placental malaria / VAR2CSA / Duffy binding domain / CSA / PfEMP1 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














