登録情報 データベース : EMDB / ID : EMD-22259タイトル Human 20S proteasome bound to an engineered 11S (PA26) activator Sharpened and masked 複合体 : PA26-bound proteasome複合体 : Proteasome複合体 : mutant PA26 / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
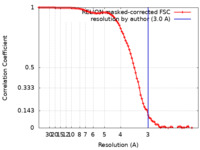
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト) / Trypanosoma brucei (トリパノソーマ)手法 / / 解像度 : 3.0 Å de la Pena AH / Opoku-Nsiah KA / Williams SK / Chopra N / Sali A / Gestwicki JE / Lander GC 資金援助 Organization Grant number 国 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) DP2-EB020402 American Cancer Society 132279-PF-18-189-01- DMC
ジャーナル : Nat Commun / 年 : 2022タイトル : The YΦ motif defines the structure-activity relationships of human 20S proteasome activators.著者 : Kwadwo A Opoku-Nsiah / Andres H de la Pena / Sarah K Williams / Nikita Chopra / Andrej Sali / Gabriel C Lander / Jason E Gestwicki / 要旨 : The 20S proteasome (20S) facilitates turnover of most eukaryotic proteins. Substrate entry into the 20S first requires opening of gating loops through binding of HbYX motifs that are present at the C- ... The 20S proteasome (20S) facilitates turnover of most eukaryotic proteins. Substrate entry into the 20S first requires opening of gating loops through binding of HbYX motifs that are present at the C-termini of certain proteasome activators (PAs). The HbYX motif has been predominantly characterized in the archaeal 20S, whereas little is known about the sequence preferences of the human 20S (h20S). Here, we synthesize and screen ~120 HbYX-like peptides, revealing unexpected differences from the archaeal system and defining the h20S recognition sequence as the Y-F/Y (YФ) motif. To gain further insight, we create a functional chimera of the optimized sequence, NLSYYT, fused to the model activator, PA26. A cryo-EM structure of PA26-h20S is used to identify key interactions, including non-canonical contacts and gate-opening mechanisms. Finally, we demonstrate that the YФ sequence preferences are tuned by valency, allowing multivalent PAs to sample greater sequence space. These results expand the model for termini-mediated gating and provide a template for the design of h20S activators. 履歴 登録 2020年6月30日 - ヘッダ(付随情報) 公開 2020年7月22日 - マップ公開 2020年7月22日 - 更新 2024年5月15日 - 現状 2024年5月15日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト) /
Homo sapiens (ヒト) / 
 データ登録者
データ登録者 米国, 2件
米国, 2件  引用
引用 ジャーナル: Nat Commun / 年: 2022
ジャーナル: Nat Commun / 年: 2022
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_22259.map.gz
emd_22259.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-22259-v30.xml
emd-22259-v30.xml emd-22259.xml
emd-22259.xml EMDBヘッダ
EMDBヘッダ emd_22259_fsc.xml
emd_22259_fsc.xml FSCデータファイル
FSCデータファイル emd_22259.png
emd_22259.png emd_22259_msk_1.map
emd_22259_msk_1.map マスクマップ
マスクマップ emd-22259.cif.gz
emd-22259.cif.gz emd_22259_half_map_1.map.gz
emd_22259_half_map_1.map.gz emd_22259_half_map_2.map.gz
emd_22259_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-22259
http://ftp.pdbj.org/pub/emdb/structures/EMD-22259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22259
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22259 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_22259.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_22259.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_22259_msk_1.map
emd_22259_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)