+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2193 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
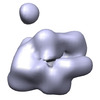
| Title | human RNA polymerase II in complex with B1 RNA | |||||||||
 Map data Map data | human RNA polymerase II in complex with B1 RNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription regulation / RNA polymerase II / non-coding RNA | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 25.0 Å | |||||||||
 Authors Authors | Kassube SA / Fang J / Grob P / Yakovchuk P / Goodrich JA / Nogales E | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2013 Journal: J Mol Biol / Year: 2013Title: Structural insights into transcriptional repression by noncoding RNAs that bind to human Pol II. Authors: Susanne A Kassube / Jie Fang / Patricia Grob / Petro Yakovchuk / James A Goodrich / Eva Nogales /  Abstract: Gene transcription is regulated in response to environmental changes and developmental cues. In mammalian cells subjected to stress conditions such as heat shock, transcription of most protein-coding ...Gene transcription is regulated in response to environmental changes and developmental cues. In mammalian cells subjected to stress conditions such as heat shock, transcription of most protein-coding genes decreases, while the transcription of heat shock protein genes increases. Repression involves direct binding to RNA polymerase II (Pol II) of certain noncoding RNAs (ncRNAs) that are upregulated upon heat shock. Another class of ncRNAs is also upregulated and binds to Pol II but does not inhibit transcription. Incorporation of repressive ncRNAs into pre-initiation complexes prevents transcription initiation, while non-repressive ncRNAs are displaced from Pol II by TFIIF. Here, we present cryo-electron microscopy reconstructions of human Pol II in complex with six different ncRNAs from mouse and human. Our structures show that both repressive and non-repressive ncRNAs bind to a conserved binding site within the cleft of Pol II. The site, which is also shared with a previously characterized yeast aptamer, is close to the active center and, thus, in an ideal position to regulate transcription. Importantly, additional RNA elements extend flexibly beyond the docking site. We propose that the differences concerning the repressive activity of the ncRNAs analyzed must be due to the distinct character of these more unstructured, flexible segments of the RNA that emanate from the cleft. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2193.map.gz emd_2193.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2193-v30.xml emd-2193-v30.xml emd-2193.xml emd-2193.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2193.png emd_2193.png | 66.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2193 http://ftp.pdbj.org/pub/emdb/structures/EMD-2193 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2193 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2193 | HTTPS FTP |
-Validation report
| Summary document |  emd_2193_validation.pdf.gz emd_2193_validation.pdf.gz | 189.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2193_full_validation.pdf.gz emd_2193_full_validation.pdf.gz | 188.3 KB | Display | |
| Data in XML |  emd_2193_validation.xml.gz emd_2193_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2193 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2193 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2193 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2193 | HTTPS FTP |
-Related structure data
| Related structure data |  2188C  2189C  2190C  2191C  2192C  2194C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2193.map.gz / Format: CCP4 / Size: 11.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2193.map.gz / Format: CCP4 / Size: 11.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human RNA polymerase II in complex with B1 RNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.54 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : human RNA polymerase II in complex with B1 RNA
+Supramolecule #1000: human RNA polymerase II in complex with B1 RNA
+Macromolecule #1: Rpb12
+Macromolecule #3: Rpb11
+Macromolecule #4: Rpb10
+Macromolecule #5: Rpb9
+Macromolecule #6: Rpb8
+Macromolecule #7: Rpb7
+Macromolecule #8: Rpb6
+Macromolecule #9: Rpb5
+Macromolecule #10: Rpb4
+Macromolecule #11: Rpb3
+Macromolecule #12: Rpb2
+Macromolecule #13: Rpb1
+Macromolecule #2: B1 RNA
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0312 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 10 mM Tris pH 7.9, 10 mM HEPES pH 8.0, 4 mM MgCl2, 50 mM KCl, 0.05% NP-40, 1 mM DTT, 0.1% trehalose |
| Grid | Details: 400 mesh copper grids covered with a holey carbon film, thin carbon film floated on top |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 86 K / Instrument: FEI VITROBOT MARK II / Method: 6 sec blot, offset -2 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Temperature | Average: 78 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Date | Oct 6, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 12.7 µm / Number real images: 133 / Average electron dose: 20 e/Å2 / Od range: 1 / Bits/pixel: 14 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50280 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side-entry cryostage / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: whole micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2, FREALIGN Details: Projection matching using a low-pass filtered model of apo Pol II as the starting model Number images used: 25079 |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















