+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21896 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full-length human ENaC ECD | |||||||||||||||
 Map data Map data | ENaC_FL sharpened EM map (Bfactor -101.4) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | sodium channel / blood pressure / epithelial / salt transport / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of salty taste / Sensory perception of salty taste / sensory perception of sour taste / neutrophil-mediated killing of bacterium / aldosterone metabolic process / leukocyte activation involved in inflammatory response / cellular response to vasopressin / sperm principal piece / sodium channel complex / epithelial fluid transport ...sensory perception of salty taste / Sensory perception of salty taste / sensory perception of sour taste / neutrophil-mediated killing of bacterium / aldosterone metabolic process / leukocyte activation involved in inflammatory response / cellular response to vasopressin / sperm principal piece / sodium channel complex / epithelial fluid transport / mucus secretion / sodium ion homeostasis / cellular response to aldosterone / renal system process / artery smooth muscle contraction / neutrophil activation involved in immune response / multicellular organismal-level water homeostasis / potassium ion homeostasis / intracellular sodium ion homeostasis / cellular response to acidic pH / sodium ion import across plasma membrane / ligand-gated sodium channel activity / motile cilium / response to food / erythrocyte homeostasis / ciliary membrane / WW domain binding / monoatomic ion channel activity / sodium ion transmembrane transport / cytoplasmic vesicle membrane / acrosomal vesicle / regulation of blood pressure / Stimuli-sensing channels / multicellular organism growth / gene expression / apical plasma membrane / response to xenobiotic stimulus / external side of plasma membrane / extracellular exosome / nucleoplasm / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||
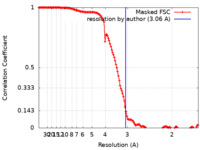
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||||||||
 Authors Authors | Posert R / Baconguis I / Noreng S / Bharadwaj A / Houser A | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Molecular principles of assembly, activation, and inhibition in epithelial sodium channel. Authors: Sigrid Noreng / Richard Posert / Arpita Bharadwaj / Alexandra Houser / Isabelle Baconguis /  Abstract: The molecular bases of heteromeric assembly and link between Na self-inhibition and protease-sensitivity in epithelial sodium channels (ENaCs) are not fully understood. Previously, we demonstrated ...The molecular bases of heteromeric assembly and link between Na self-inhibition and protease-sensitivity in epithelial sodium channels (ENaCs) are not fully understood. Previously, we demonstrated that ENaC subunits - α, β, and γ - assemble in a counterclockwise configuration when viewed from outside the cell with the protease-sensitive GRIP domains in the periphery (Noreng et al., 2018). Here we describe the structure of ENaC resolved by cryo-electron microscopy at 3 Å. We find that a combination of precise domain arrangement and complementary hydrogen bonding network defines the subunit arrangement. Furthermore, we determined that the α subunit has a primary functional module consisting of the finger and GRIP domains. The module is bifurcated by the α2 helix dividing two distinct regulatory sites: Na and the inhibitory peptide. Removal of the inhibitory peptide perturbs the Na site via the α2 helix highlighting the critical role of the α2 helix in regulating ENaC function. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21896.map.gz emd_21896.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21896-v30.xml emd-21896-v30.xml emd-21896.xml emd-21896.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21896_fsc.xml emd_21896_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_21896.png emd_21896.png | 88.7 KB | ||
| Masks |  emd_21896_msk_1.map emd_21896_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21896.cif.gz emd-21896.cif.gz | 7.3 KB | ||
| Others |  emd_21896_additional.map.gz emd_21896_additional.map.gz emd_21896_half_map_1.map.gz emd_21896_half_map_1.map.gz emd_21896_half_map_2.map.gz emd_21896_half_map_2.map.gz | 108.1 MB 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21896 http://ftp.pdbj.org/pub/emdb/structures/EMD-21896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21896 | HTTPS FTP |
-Validation report
| Summary document |  emd_21896_validation.pdf.gz emd_21896_validation.pdf.gz | 996 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21896_full_validation.pdf.gz emd_21896_full_validation.pdf.gz | 995.5 KB | Display | |
| Data in XML |  emd_21896_validation.xml.gz emd_21896_validation.xml.gz | 21.8 KB | Display | |
| Data in CIF |  emd_21896_validation.cif.gz emd_21896_validation.cif.gz | 28.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21896 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21896 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21896 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21896 | HTTPS FTP |
-Related structure data
| Related structure data |  6wthMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21896.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21896.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ENaC_FL sharpened EM map (Bfactor -101.4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
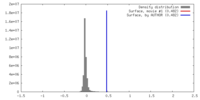
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21896_msk_1.map emd_21896_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
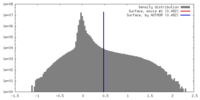
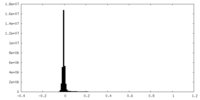
| Density Histograms |
-Additional map: ENaC FL unsharpened EM map
| File | emd_21896_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ENaC_FL unsharpened EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
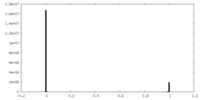
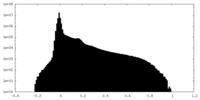
| Density Histograms |
-Half map: ENaC FL EM half map A
| File | emd_21896_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ENaC_FL EM half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
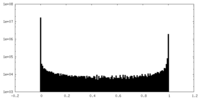
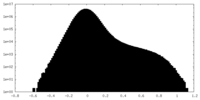
| Density Histograms |
-Half map: ENaC FL EM half map B
| File | emd_21896_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ENaC_FL EM half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
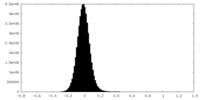
| Density Histograms |
- Sample components
Sample components
-Entire : Full-length human ENaC heterotrimer bound with two high-affinity Fabs
| Entire | Name: Full-length human ENaC heterotrimer bound with two high-affinity Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Full-length human ENaC heterotrimer bound with two high-affinity Fabs
| Supramolecule | Name: Full-length human ENaC heterotrimer bound with two high-affinity Fabs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 320 KDa |
-Macromolecule #1: Amiloride-sensitive sodium channel subunit alpha
| Macromolecule | Name: Amiloride-sensitive sodium channel subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.780531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGNKLEEQD SSPPQSTPGL MKGNKREEQG LGPEPAAPQQ PTAEEEALIE FHRSYRELFE FFCNNTTIHG AIRLVCSQHN RMKTAFWAV LWLCTFGMMY WQFGLLFGEY FSYPVSLNIN LNSDKLVFPA VTICTLNPYR YPEIKEELEE LDRITEQTLF D LYKYSSFT ...String: MEGNKLEEQD SSPPQSTPGL MKGNKREEQG LGPEPAAPQQ PTAEEEALIE FHRSYRELFE FFCNNTTIHG AIRLVCSQHN RMKTAFWAV LWLCTFGMMY WQFGLLFGEY FSYPVSLNIN LNSDKLVFPA VTICTLNPYR YPEIKEELEE LDRITEQTLF D LYKYSSFT TLVAGSRSRR DLRGTLPHPL QRLRVPPPPH GARRARSVAS SLRDNNPQVD WKDWKIGFQL CNQNKSDCFY QT YSSGVDA VREWYRFHYI NILSRLPETL PSLEEDTLGN FIFACRFNQV SCNQANYSHF HHPMYGNCYT FNDKNNSNLW MSS MPGINN GLSLMLRAEQ NDFIPLLSTV TGARVMVHGQ DEPAFMDDGG FNLRPGVETS ISMRKETLDR LGGDYGDCTK NGSD VPVEN LYPSKYTQQV CIHSCFQESM IKECGCAYIF YPRPQNVEYC DYRKHSSWGY CYYKLQVDFS SDHLGCFTKC RKPCS VTSY QLSAGYSRWP SVTSQEWVFQ MLSRQNNYTV NNKRNGVAKV NIFFKELNYK TNSESPSVTM VTLLSNLGSQ WSLWFG SSV LSVVEMAELV FDLLVIMFLM LLRRFRSRYW SPGRGGRGAQ EVASTLASSP PSHFCPHPMS LSLSQPGPAP SPALTAP PP AYATLGPRPS PGGSAGASSS TCPLGGP UniProtKB: Epithelial sodium channel subunit alpha |
-Macromolecule #2: Amiloride-sensitive sodium channel subunit beta
| Macromolecule | Name: Amiloride-sensitive sodium channel subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.728891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHVKKYLLKG LHRLQKGPGY TYKELLVWYC DNTNTHGPKR IICEGPKKKA MWFLLTLLFA ALVCWQWGIF IRTYLSWEVS VSLSVGFKT MDFPAVTICN ASPFKYSKIK HLLKDLDELM EAVLERILAP ELSHANATRN LNFSIWNHTP LVLIDERNPH H PMVLDLFG ...String: MHVKKYLLKG LHRLQKGPGY TYKELLVWYC DNTNTHGPKR IICEGPKKKA MWFLLTLLFA ALVCWQWGIF IRTYLSWEVS VSLSVGFKT MDFPAVTICN ASPFKYSKIK HLLKDLDELM EAVLERILAP ELSHANATRN LNFSIWNHTP LVLIDERNPH H PMVLDLFG DNHNGLTSSS ASEKICNAHG CKMAMRLCSL NRTQCTFRNF TSATQALTEW YILQATNIFA QVPQQELVEM SY PGEQMIL ACLFGAEPCN YRNFTSIFYP HYGNCYIFNW GMTEKALPSA NPGTEFGLKL ILDIGQEDYV PFLASTAGVR LML HEQRSY PFIRDEGIYA MSGTETSIGV LVDKLQRMGE PYSPCTVNGS EVPVQNFYSD YNTTYSIQAC LRSCFQDHMI RNCN CGHYL YPLPRGEKYC NNRDFPDWAH CYSDLQMSVA QRETCIGMCK ESCNDTQYKM TISMADWPSE ASEDWIFHVL SQERD QSTN ITLSRKGIVK LNIYFQEFNY RTIEESAANN IVWLLSNLGG QFGFWMGGSV LCLIEFGEII IDFVWITIIK LVALAK SLR QRRAQASYAG PPPTVAELVE AHTNFGFQPD TAPRSPNTGP YPSEQALPIP GTPPPNYDSL RLQPLDVIES DSEGDAI UniProtKB: Epithelial sodium channel subunit beta |
-Macromolecule #3: Amiloride-sensitive sodium channel subunit gamma
| Macromolecule | Name: Amiloride-sensitive sodium channel subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.352984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAPGEKIKAK IKKNLPVTGP QAPTIKELMR WYCLNTNTHG CRRIVVSRGR LRRLLWIGFT LTAVALILWQ CALLVFSFYT VSVSIKVHF RKLDFPAVTI CNINPYKYST VRHLLADLEQ ETREALKSLY GFPESRKRRE AESWNSVSEG KQPRFSHRIP L LIFDQDEK ...String: MAPGEKIKAK IKKNLPVTGP QAPTIKELMR WYCLNTNTHG CRRIVVSRGR LRRLLWIGFT LTAVALILWQ CALLVFSFYT VSVSIKVHF RKLDFPAVTI CNINPYKYST VRHLLADLEQ ETREALKSLY GFPESRKRRE AESWNSVSEG KQPRFSHRIP L LIFDQDEK GKARDFFTGR KRKVGGSIIH KASNVMHIES KQVVGFQLCS NDTSDCATYT FSSGINAIQE WYKLHYMNIM AQ VPLEKKI NMSYSAEELL VTCFFDGVSC DARNFTLFHH PMHGNCYTFN NRENETILST SMGGSEYGLQ VILYINEEEY NPF LVSSTG AKVIIHRQDE YPFVEDVGTE IETAMVTSIG MHLTESFKLS EPYSQCTEDG SDVPIRNIYN AAYSLQICLH SCFQ TKMVE KCGCAQYSQP LPPAANYCNY QQHPNWMYCY YQLHRAFVQE ELGCQSVCKE ACSFKEWTLT TSLAQWPSVV SEKWL LPVL TWDQGRQVNK KLNKTDLAKL LIFYKDLNQR SIMESPANSI EMLLSNFGGQ LGLWMSCSVV CVIEIIEVFF IDFFSI IAR RQWQKAKEWW AWKQAPPCPE APRSPQGQDN PALDIDDDLP TFNSALHLPP ALGTQVPGTP PPKYNTLRLE RAFSNQL TD TQMLDEL UniProtKB: Epithelial sodium channel subunit gamma |
-Macromolecule #4: 7B1 Fab
| Macromolecule | Name: 7B1 Fab / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.060393 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #5: 10D4 Fab
| Macromolecule | Name: 10D4 Fab / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.805078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK) |
-Macromolecule #7: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 7 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK III Details: 3.5 uL applied, manual blot, fresh 3.5 uL applied, vitrobot blot and freeze. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)