+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2172 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Methanococcus igneus 70S ribosome | |||||||||
 Map data Map data | Methanococcus igneus 70S ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | archaea / ribosome 70S protein / RNA / kink-turn / protein synthesis mass spectrometry | |||||||||
| Biological species |  Methanotorris igneus (archaea) Methanotorris igneus (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 18.0 Å | |||||||||
 Authors Authors | Armache J-P / Anger AM / Marquez V / Frankenberg S / Froehlich T / Villa E / Berninghausen O / Thomm M / Arnold GJ / Beckmann R / Wilson DN | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2013 Journal: Nucleic Acids Res / Year: 2013Title: Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Authors: Jean-Paul Armache / Andreas M Anger / Viter Márquez / Sibylle Franckenberg / Thomas Fröhlich / Elizabeth Villa / Otto Berninghausen / Michael Thomm / Georg J Arnold / Roland Beckmann / Daniel N Wilson /  Abstract: In all living cells, protein synthesis occurs on ribonucleoprotein particles called ribosomes. Molecular models have been reported for complete bacterial 70S and eukaryotic 80S ribosomes; however, ...In all living cells, protein synthesis occurs on ribonucleoprotein particles called ribosomes. Molecular models have been reported for complete bacterial 70S and eukaryotic 80S ribosomes; however, only molecular models of large 50S subunits have been reported for archaea. Here, we present a complete molecular model for the Pyrococcus furiosus 70S ribosome based on a 6.6 Å cryo-electron microscopy map. Moreover, we have determined cryo-electron microscopy reconstructions of the Euryarchaeota Methanococcus igneus and Thermococcus kodakaraensis 70S ribosomes and Crenarchaeota Staphylothermus marinus 50S subunit. Examination of these structures reveals a surprising promiscuous behavior of archaeal ribosomal proteins: We observe intersubunit promiscuity of S24e and L8e (L7ae), the latter binding to the head of the small subunit, analogous to S12e in eukaryotes. Moreover, L8e and L14e exhibit intrasubunit promiscuity, being present in two copies per archaeal 50S subunit, with the additional binding site of L14e analogous to the related eukaryotic r-protein L27e. Collectively, these findings suggest insights into the evolution of eukaryotic ribosomal proteins through increased copy number and binding site promiscuity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2172.map.gz emd_2172.map.gz | 17.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2172-v30.xml emd-2172-v30.xml emd-2172.xml emd-2172.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2172.png EMD-2172.png | 132.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2172 http://ftp.pdbj.org/pub/emdb/structures/EMD-2172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2172 | HTTPS FTP |
-Validation report
| Summary document |  emd_2172_validation.pdf.gz emd_2172_validation.pdf.gz | 234.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2172_full_validation.pdf.gz emd_2172_full_validation.pdf.gz | 233.5 KB | Display | |
| Data in XML |  emd_2172_validation.xml.gz emd_2172_validation.xml.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2172 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2172 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2172 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2172 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2172.map.gz / Format: CCP4 / Size: 185.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2172.map.gz / Format: CCP4 / Size: 185.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Methanococcus igneus 70S ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
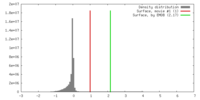
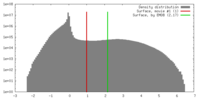
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Methanococcus igneus 70S ribosome
| Entire | Name: Methanococcus igneus 70S ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: Methanococcus igneus 70S ribosome
| Supramolecule | Name: Methanococcus igneus 70S ribosome / type: sample / ID: 1000 / Oligomeric state: One ribosome / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 2.1 MDa / Theoretical: 2.1 MDa |
-Supramolecule #1: Methanococcus igneus 70S ribosome
| Supramolecule | Name: Methanococcus igneus 70S ribosome / type: complex / ID: 1 / Name.synonym: 70S ribosome / Recombinant expression: No Ribosome-details: ribosome-prokaryote: LSU 50S, LSU RNA 23S, LSU RNA 5S, SSU 30S, PSR16s, ALL |
|---|---|
| Source (natural) | Organism:  Methanotorris igneus (archaea) Methanotorris igneus (archaea) |
| Molecular weight | Experimental: 2.1 MDa / Theoretical: 2.1 MDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Staining | Type: NEGATIVE Details: Blot for 10 seconds before plunging, use 2 layers of filter paper |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III / Details: Vitrification instrument: Vitrobot Method: Blot for 10 seconds before plunging, use 2 layers of filter paper |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Date | Jul 9, 2008 |
| Image recording | Category: FILM / Film or detector model: FEI EAGLE (4k x 4k) / Digitization - Scanner: OTHER / Number real images: 99 / Average electron dose: 20 e/Å2 / Details: Data collected on CCD |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Wiener filter on 3D volumes |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 8932 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Details | Protocol: Rigid-body fit into the density using UCSF Chimera built-in "Fit in Map". Rigid-body fit into the density using UCSF Chimera built-in "Fit in Map" |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)