[English] 日本語
 Yorodumi
Yorodumi- EMDB-21706: Cryogenic Single-Molecule Fluorescence Annotations for Electron T... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21706 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryogenic Single-Molecule Fluorescence Annotations for Electron Tomography Reveal In Situ Organization of Key Proteins in Caulobacter | |||||||||||||||
 Map data Map data | Tomographic Reconstruction (bin by 4) of C. Crescentus that underwent no correlative imaging. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Bacteria / Tomography / Regulatory proteins / CELL CYCLE | |||||||||||||||
| Biological species |  Caulobacter vibrioides (bacteria) Caulobacter vibrioides (bacteria) | |||||||||||||||
| Method | electron tomography / cryo EM | |||||||||||||||
 Authors Authors | Dahlberg PD / Saurabh S / Sartor AM / Wang J / Mitchell P / Chiu W / Shapiro L / Moerner WE | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Cryogenic single-molecule fluorescence annotations for electron tomography reveal in situ organization of key proteins in . Authors: Peter D Dahlberg / Saumya Saurabh / Annina M Sartor / Jiarui Wang / Patrick G Mitchell / Wah Chiu / Lucy Shapiro / W E Moerner /  Abstract: Superresolution fluorescence microscopy and cryogenic electron tomography (CET) are powerful imaging methods for exploring the subcellular organization of biomolecules. Superresolution fluorescence ...Superresolution fluorescence microscopy and cryogenic electron tomography (CET) are powerful imaging methods for exploring the subcellular organization of biomolecules. Superresolution fluorescence microscopy based on covalent labeling highlights specific proteins and has sufficient sensitivity to observe single fluorescent molecules, but the reconstructions lack detailed cellular context. CET has molecular-scale resolution but lacks specific and nonperturbative intracellular labeling techniques. Here, we describe an imaging scheme that correlates cryogenic single-molecule fluorescence localizations with CET reconstructions. Our approach achieves single-molecule localizations with an average lateral precision of 9 nm, and a relative registration error between the set of localizations and CET reconstruction of ∼30 nm. We illustrate the workflow by annotating the positions of three proteins in the bacterium : McpA, PopZ, and SpmX. McpA, which forms a part of the chemoreceptor array, acts as a validation structure by being visible under both imaging modalities. In contrast, PopZ and SpmX cannot be directly identified in CET. While not directly discernable, PopZ fills a region at the cell poles that is devoid of electron-dense ribosomes. We annotate the position of PopZ with single-molecule localizations and confirm its position within the ribosome excluded region. We further use the locations of PopZ to provide context for localizations of SpmX, a low-copy integral membrane protein sequestered by PopZ as part of a signaling pathway that leads to an asymmetric cell division. Our correlative approach reveals that SpmX localizes along one side of the cell pole and its extent closely matches that of the PopZ region. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21706.map.gz emd_21706.map.gz | 130.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21706-v30.xml emd-21706-v30.xml emd-21706.xml emd-21706.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21706.png emd_21706.png | 97.6 KB | ||
| Others |  emd_21706_additional_1.map.gz emd_21706_additional_1.map.gz emd_21706_additional_2.map.gz emd_21706_additional_2.map.gz emd_21706_additional_3.map.gz emd_21706_additional_3.map.gz emd_21706_additional_4.map.gz emd_21706_additional_4.map.gz emd_21706_additional_5.map.gz emd_21706_additional_5.map.gz | 543.6 MB 589.7 KB 178.8 MB 774.5 KB 118.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21706 http://ftp.pdbj.org/pub/emdb/structures/EMD-21706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21706 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21706.map.gz / Format: CCP4 / Size: 590.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21706.map.gz / Format: CCP4 / Size: 590.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomographic Reconstruction (bin by 4) of C. Crescentus that underwent no correlative imaging. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 29.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
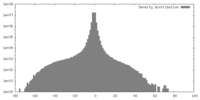
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Tomographic Reconstruction (bin 4) of C. Crescentus that...
| File | emd_21706_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomographic Reconstruction (bin 4) of C. Crescentus that underwent correlative imaging. See corresponding file for single-molecule fluorescence localizations annotating the positions of PAmKate-PopZ. | ||||||||||||
| Projections & Slices |
| ||||||||||||
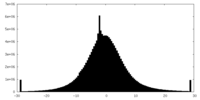
| Density Histograms |
-Additional map: Single-molecule fluorescence localizations annotating the positions of PAmKate-PopZ....
| File | emd_21706_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-molecule fluorescence localizations annotating the positions of PAmKate-PopZ. | ||||||||||||
| Projections & Slices |
| ||||||||||||
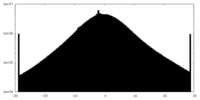
| Density Histograms |
-Additional map: Tomographic Reconstruction (bin 4) of C. Crescentus that...
| File | emd_21706_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomographic Reconstruction (bin 4) of C. Crescentus that underwent correlative imaging. See corresponding file for single-molecule fluorescence localizations annotating the positions of PAmKate-McpA. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Single-molecule fluorescence localizations annotating the positions of PAmKate-McpA....
| File | emd_21706_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-molecule fluorescence localizations annotating the positions of PAmKate-McpA. | ||||||||||||
| Projections & Slices |
| ||||||||||||
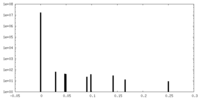
| Density Histograms |
-Additional map: Tomographic Reconstruction (bin by 4) of C. Crescentus...
| File | emd_21706_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomographic Reconstruction (bin by 4) of C. Crescentus that underwent correlative imaging. Fluorescence imaging was performed using an excitation beam with an intensity of ~80W/cm^2 for ~3 hours. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Caulobacter crescentus cells
| Entire | Name: Caulobacter crescentus cells |
|---|---|
| Components |
|
-Supramolecule #1: Caulobacter crescentus cells
| Supramolecule | Name: Caulobacter crescentus cells / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides (bacteria) Caulobacter vibrioides (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 / Details: Cells cultured and vitrified in M2G growth media |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 / Details: double sided blot for 3.5 seconds. |
| Cryo protectant | 20% ethylene glycol |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: EMS / Diameter: 15 nm |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 1.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 10.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 91 IMOD / Number images used: 91 |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)