+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21672 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PI3P and calcium bound full-length TRPY1 in detergent | |||||||||
 Map data Map data | final post-processed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTRP channels / vacuole-mitochondrion membrane contact site / intracellular monoatomic cation homeostasis / voltage-gated monoatomic ion channel activity / fungal-type vacuole / calcium-activated cation channel activity / fungal-type vacuole membrane / calcium ion transmembrane import into cytosol / sodium channel activity / calcium ion import across plasma membrane ...TRP channels / vacuole-mitochondrion membrane contact site / intracellular monoatomic cation homeostasis / voltage-gated monoatomic ion channel activity / fungal-type vacuole / calcium-activated cation channel activity / fungal-type vacuole membrane / calcium ion transmembrane import into cytosol / sodium channel activity / calcium ion import across plasma membrane / potassium channel activity / calcium channel activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
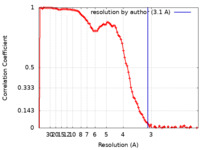
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Ahmed T / Moiseenkova-Bell VY | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structure of the ancient TRPY1 channel from Saccharomyces cerevisiae reveals mechanisms of modulation by lipids and calcium. Authors: Tofayel Ahmed / Collin R Nisler / Edwin C Fluck / Sanket Walujkar / Marcos Sotomayor / Vera Y Moiseenkova-Bell /  Abstract: Transient receptor potential (TRP) channels emerged in fungi as mechanosensitive osmoregulators. The Saccharomyces cerevisiae vacuolar TRP yeast 1 (TRPY1) is the most studied TRP channel from fungi, ...Transient receptor potential (TRP) channels emerged in fungi as mechanosensitive osmoregulators. The Saccharomyces cerevisiae vacuolar TRP yeast 1 (TRPY1) is the most studied TRP channel from fungi, but the structure and details of channel modulation remain elusive. Here, we describe the full-length cryoelectron microscopy structure of TRPY1 at 3.1 Å resolution in a closed state. The structure, despite containing an evolutionarily conserved and archetypical transmembrane domain, reveals distinctive structural folds for the cytosolic N and C termini, compared with other eukaryotic TRP channels. We identify an inhibitory phosphatidylinositol 3-phosphate (PI(3)P) lipid-binding site, along with two Ca-binding sites: a cytosolic site, implicated in channel activation and a vacuolar lumen site, implicated in inhibition. These findings, together with data from microsecond-long molecular dynamics simulations and a model of a TRPY1 open state, provide insights into the basis of TRPY1 channel modulation by lipids and Ca, and the molecular evolution of TRP channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21672.map.gz emd_21672.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21672-v30.xml emd-21672-v30.xml emd-21672.xml emd-21672.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21672_fsc.xml emd_21672_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21672.png emd_21672.png | 104 KB | ||
| Masks |  emd_21672_msk_1.map emd_21672_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21672.cif.gz emd-21672.cif.gz | 5.7 KB | ||
| Others |  emd_21672_half_map_1.map.gz emd_21672_half_map_1.map.gz emd_21672_half_map_2.map.gz emd_21672_half_map_2.map.gz | 45.7 MB 45.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21672 http://ftp.pdbj.org/pub/emdb/structures/EMD-21672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21672 | HTTPS FTP |
-Related structure data
| Related structure data |  6whgMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21672.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21672.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21672_msk_1.map emd_21672_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map2
| File | emd_21672_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map1
| File | emd_21672_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PI3P and calcium bound full-length TRPY1 in detergent
| Entire | Name: PI3P and calcium bound full-length TRPY1 in detergent |
|---|---|
| Components |
|
-Supramolecule #1: PI3P and calcium bound full-length TRPY1 in detergent
| Supramolecule | Name: PI3P and calcium bound full-length TRPY1 in detergent / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Calcium channel YVC1
| Macromolecule | Name: Calcium channel YVC1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.034602 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVSANGDLHL PISNEQCMPE NNGSLGFEAP TPRQILRVTL NLKYLIDKVV PIVYDPNDIV CDHSEILSPK VVKLAYEACG GNPKDKANK RKYQSVIIFS LLKVCEWYSI LATMEVHNAK LYETRNLASQ QLCKLLIERE ETRDLQFLFM QLLLRRYVIN E NDEDQEPL ...String: MVSANGDLHL PISNEQCMPE NNGSLGFEAP TPRQILRVTL NLKYLIDKVV PIVYDPNDIV CDHSEILSPK VVKLAYEACG GNPKDKANK RKYQSVIIFS LLKVCEWYSI LATMEVHNAK LYETRNLASQ QLCKLLIERE ETRDLQFLFM QLLLRRYVIN E NDEDQEPL NALELATDMH CTTVIGSSGF QRCLKWIWRG WIVQNGLDPT TFIKDDSLAE VSLISHFNPV RLKAPVYQNY LQ MIFSFLF LGLYTLVVNG KDSERVQSFD LLESIFYVFN TGFILDELTK LYYIGYAHLS FWNLFNDTTY LIITFAMGFR AMS VTPLNA KYSSEDWDKI SYRVLSCAAP FVWSRLLLYL ESQRFIGIML VILKHMMKES IVFFFLLFLI MIGFTQGFLG LDSA DGKRD ITGPILGNLT ITVLGLGSFD VFEEFAPPYA AILYYGYYFI VSVILLNILI ALYSTAYQKV IDNADDEYMA LMSQK TLRY IRAPDEDVYV SPLNLIEVFM TPIFRILPPK RAKDLSYTVM TIVYSPFLLL ISVKETREAR RIKYNRMKRL NDDANE YDT PWDLTDGYLD DDDGLFSDNR NSGMRATQLK NSRSLKLQRT AEQEDVHFKV PKKWYKNVKK CSPSFEQYDN DDTEDDA GE DKDEVKELTK KVENLTAVIT DLLEKLDIKD KKE UniProtKB: Calcium channel YVC1 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 8 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: (2R)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2S,3S,4S,5S,6R)-2,3,4,6...
| Macromolecule | Name: (2R)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2S,3S,4S,5S,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl hexadecanoate type: ligand / ID: 3 / Number of copies: 4 / Formula: PWE |
|---|---|
| Molecular weight | Theoretical: 722.693 Da |
| Chemical component information |  ChemComp-PWE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)