[English] 日本語
 Yorodumi
Yorodumi- EMDB-21456: Cryo-EM structure of M1214_N1 Fab in complex with CH505 TF chimer... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21456 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer | ||||||||||||
 Map data Map data | M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cryo-EM / broadly neutralizing antibody / bNAb / Fab / human immunodeficiency virus / HIV-1 / CH505 / SOSIP / Env / IMMUNE SYSTEM | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.86 Å | ||||||||||||
 Authors Authors | Chan K-W / Kong XP | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2020 Journal: Cell Host Microbe / Year: 2020Title: VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA. Authors: Manxue Jia / Rachel A Liberatore / Yicheng Guo / Kun-Wei Chan / Ruimin Pan / Hong Lu / Eric Waltari / Eva Mittler / Kartik Chandran / Andrés Finzi / Daniel E Kaufmann / Michael S Seaman / ...Authors: Manxue Jia / Rachel A Liberatore / Yicheng Guo / Kun-Wei Chan / Ruimin Pan / Hong Lu / Eric Waltari / Eva Mittler / Kartik Chandran / Andrés Finzi / Daniel E Kaufmann / Michael S Seaman / David D Ho / Lawrence Shapiro / Zizhang Sheng / Xiang-Peng Kong / Paul D Bieniasz / Xueling Wu /   Abstract: The HIV-1 envelope (Env) undergoes conformational changes during infection. Broadly neutralizing antibodies (bNAbs) are typically isolated by using soluble Env trimers, which do not capture all Env ...The HIV-1 envelope (Env) undergoes conformational changes during infection. Broadly neutralizing antibodies (bNAbs) are typically isolated by using soluble Env trimers, which do not capture all Env states. To address these limitations, we devised a vesicular stomatitis virus (VSV)-based probe to display membrane-embedded Env trimers and isolated five bNAbs from two chronically infected donors, M4008 and M1214. Donor B cell receptor (BCR) repertoires identified two bNAb lineages, M4008_N1 and M1214_N1, that class-switched to immunoglobulin G (IgG) and IgA. Variants of these bNAbs reconstituted as IgA demonstrated broadly neutralizing activity, and the IgA fraction of M1214 plasma conferred neutralization. M4008_N1 epitope mapping revealed a glycan-independent V3 epitope conferring tier 2 virus neutralization. A 4.86-Å-resolution cryogenic electron microscopy (cryo-EM) structure of M1214_N1 complexed with CH505 SOSIP revealed another elongated epitope, the V2V5 corridor, extending from V2 to V5. Overall, the VSV probe identified bNAb lineages with neutralizing IgG and IgA members targeting distinct sites of HIV-1 Env vulnerability. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21456.map.gz emd_21456.map.gz | 167.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21456-v30.xml emd-21456-v30.xml emd-21456.xml emd-21456.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21456.png emd_21456.png | 38.7 KB | ||
| Filedesc metadata |  emd-21456.cif.gz emd-21456.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21456 http://ftp.pdbj.org/pub/emdb/structures/EMD-21456 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21456 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21456 | HTTPS FTP |
-Related structure data
| Related structure data |  6vy2MC  6vu2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21456.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21456.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
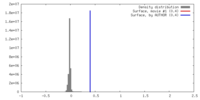
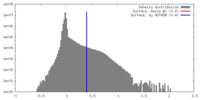
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer
| Entire | Name: M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer |
|---|---|
| Components |
|
-Supramolecule #1: M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer
| Supramolecule | Name: M1214_N1 Fab in complex with CH505 TF chimeric SOSIP.664 Env trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: chimeric CH505TF SOSIP664.R6 E64K A316W
| Supramolecule | Name: chimeric CH505TF SOSIP664.R6 E64K A316W / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: Fab fragment of the M1214_N1 IgG
| Supramolecule | Name: Fab fragment of the M1214_N1 IgG / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glycoprotein 120
| Macromolecule | Name: Glycoprotein 120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.540402 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPMGSLQPLA TLYLLGMLVA SVLAAENLWV TVYYGVPVWK EAKTTLFCAS DAKAYEKKVH NVWATHACVP TDPNPQEMVL KNVTENFNM WKNDMVDQMH EDVISLWDQS LKPCVKLTPL CVTLNCTNAT ASNSSIIEGM KNCSFNITTE LRDKREKKNA L FYKLDIVQ ...String: MPMGSLQPLA TLYLLGMLVA SVLAAENLWV TVYYGVPVWK EAKTTLFCAS DAKAYEKKVH NVWATHACVP TDPNPQEMVL KNVTENFNM WKNDMVDQMH EDVISLWDQS LKPCVKLTPL CVTLNCTNAT ASNSSIIEGM KNCSFNITTE LRDKREKKNA L FYKLDIVQ LDGNSSQYRL INCNTSVITQ ACPKVSFDPI PIHYCAPAGY AILKCNNKTF TGTGPCNNVS TVQCTHGIKP VV STQLLLN GSLAEGEIII RSENITNNVK TIIVHLNESV KIECTRPNNK TRTSIRIGPG QWFYATGQVI GDIREAYCNI NES KWNETL QRVSKKLKEY FPHKNITFQP SSGGDLEITT HSFNCGGEFF YCNTSSLFNR TYMANSTDMA NSTETNSTRT ITIH CRIKQ IINMWQEVGR AMYAPPIAGN ITCISNITGL LLTRDGGKNN TETFRPGGGN MKDNWRSELY KYKVVKIEPL GVAPT RCKR RVV UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Glycoprotein 41
| Macromolecule | Name: Glycoprotein 41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 18.146699 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GRRRRRRAVG IGAVFLGFLG AAGSTMGAAS MTLTVQARNL LSGIVQQQSN LLRAPEAQQH LLKLTVWGIK QLQARVLAVE RYLRDQQLL GIWGCSGKLI CCTNVPWNSS WSNRNLSEIW DNMTWLQWDK EISNYTQIIY GLLEESQNQQ EKNEQDLLAL D UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: M1214 N1 Fab heavy chain
| Macromolecule | Name: M1214 N1 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.210143 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DRLFQSGGGV SRPGGSLRVN CGASGFTVRT HYMYWLRQSP GKGLEWVAFM NSGGSVSYVD SVRGRFSVSR DNPANAMVLQ MDALKIEDT GTYYCARELR EAWYGDLRDY SGLDVWGRGT IVSISSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: DRLFQSGGGV SRPGGSLRVN CGASGFTVRT HYMYWLRQSP GKGLEWVAFM NSGGSVSYVD SVRGRFSVSR DNPANAMVLQ MDALKIEDT GTYYCARELR EAWYGDLRDY SGLDVWGRGT IVSISSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPK |
-Macromolecule #4: M1214 N1 Fab light chain
| Macromolecule | Name: M1214 N1 Fab light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.87642 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALAQPPSV SGSPGQSVTI TCTGINDYGA AYKFVSWYQQ HPGKEPRLIM KNVKDRWSVT PNRFSGSTSG NTASLTISNL QSDDEAQYF CAVYAGGFTF PRLGGGTKLS VLSQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT ...String: QSALAQPPSV SGSPGQSVTI TCTGINDYGA AYKFVSWYQQ HPGKEPRLIM KNVKDRWSVT PNRFSGSTSG NTASLTISNL QSDDEAQYF CAVYAGGFTF PRLGGGTKLS VLSQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT PSKQSNNKYA ASSYLSLTPE QWKSHRSYSC QVTHEGSTVE KTVAPT |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 24 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||
| Output model |  PDB-6vy2: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)