[English] 日本語
 Yorodumi
Yorodumi- EMDB-21426: Single Particle Cryo-EM Structure of the Natively Isolated Sec61 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21426 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single Particle Cryo-EM Structure of the Natively Isolated Sec61 complex, TMCO1, Nicalin, TMEM147, and CCDC47 Containing Translocon | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
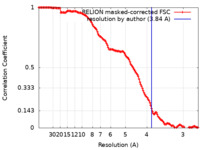
| Method | single particle reconstruction / cryo EM / Resolution: 3.84 Å | |||||||||
 Authors Authors | McGilvray PT / Anghel SA / Sundaram A / Trnka MJ / Zhong F / Hu H / Burlingame AL / Keenan RJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: An ER translocon for multi-pass membrane protein biogenesis. Authors: Philip T McGilvray / S Andrei Anghel / Arunkumar Sundaram / Frank Zhong / Michael J Trnka / James R Fuller / Hong Hu / Alma L Burlingame / Robert J Keenan /  Abstract: Membrane proteins with multiple transmembrane domains play critical roles in cell physiology, but little is known about the machinery coordinating their biogenesis at the endoplasmic reticulum. Here ...Membrane proteins with multiple transmembrane domains play critical roles in cell physiology, but little is known about the machinery coordinating their biogenesis at the endoplasmic reticulum. Here we describe a ~ 360 kDa ribosome-associated complex comprising the core Sec61 channel and five accessory factors: TMCO1, CCDC47 and the Nicalin-TMEM147-NOMO complex. Cryo-electron microscopy reveals a large assembly at the ribosome exit tunnel organized around a central membrane cavity. Similar to protein-conducting channels that facilitate movement of transmembrane segments, cytosolic and luminal funnels in TMCO1 and TMEM147, respectively, suggest routes into the central membrane cavity. High-throughput mRNA sequencing shows selective translocon engagement with hundreds of different multi-pass membrane proteins. Consistent with a role in multi-pass membrane protein biogenesis, cells lacking different accessory components show reduced levels of one such client, the glutamate transporter EAAT1. These results identify a new human translocon and provide a molecular framework for understanding its role in multi-pass membrane protein biogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21426.map.gz emd_21426.map.gz | 176 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21426-v30.xml emd-21426-v30.xml emd-21426.xml emd-21426.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21426_fsc.xml emd_21426_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_21426.png emd_21426.png | 75.3 KB | ||
| Masks |  emd_21426_msk_1.map emd_21426_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Others |  emd_21426_half_map_1.map.gz emd_21426_half_map_1.map.gz emd_21426_half_map_2.map.gz emd_21426_half_map_2.map.gz | 175.3 MB 174.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21426 http://ftp.pdbj.org/pub/emdb/structures/EMD-21426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21426 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21426.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21426.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21426_msk_1.map emd_21426_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
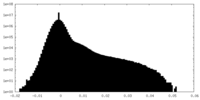
| Density Histograms |
-Half map: #1
| File | emd_21426_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
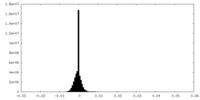
| Density Histograms |
-Half map: #2
| File | emd_21426_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
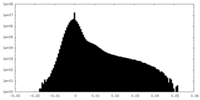
| Density Histograms |
- Sample components
Sample components
+Entire : Native complex of a translating ribosome and a novel translocon c...
+Supramolecule #1: Native complex of a translating ribosome and a novel translocon c...
+Supramolecule #2: Translocon
+Supramolecule #3: Sec61 Complex
+Supramolecule #4: TMCO1
+Supramolecule #5: NOMO-Nicalin-TMEM147 Complex
+Supramolecule #6: CCDC47
+Supramolecule #7: Sec61 alpha
+Supramolecule #8: Sec61 beta
+Supramolecule #9: Sec61 gamma
+Supramolecule #10: TMEM147
+Supramolecule #11: Nicalin
+Supramolecule #12: NOMO
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.0 nm / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: Two filter papers were added to each arm, 2.5 microliters of sample were added to grids, samples were blotted for 11 seconds, and 0.5 second of drain time was allowed before vitrification.. | |||||||||||||||
| Details | Sample was well-dispersed on a thin (~2 nm) carbon film. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 5562 / Average exposure time: 3.8 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)