[English] 日本語
 Yorodumi
Yorodumi- EMDB-21368: Cryo-EM structure of the wild-type human serotonin transporter co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21368 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the wild-type human serotonin transporter complexed with paroxetine and 8B6 Fab | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antidepressant / complex / transporter / antibody / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of cerebellar granule cell precursor proliferation / regulation of thalamus size / Serotonin clearance from the synaptic cleft / serotonergic synapse / positive regulation of serotonin secretion / negative regulation of synaptic transmission, dopaminergic / cocaine binding / serotonin:sodium:chloride symporter activity / cellular response to cGMP / enteric nervous system development ...negative regulation of cerebellar granule cell precursor proliferation / regulation of thalamus size / Serotonin clearance from the synaptic cleft / serotonergic synapse / positive regulation of serotonin secretion / negative regulation of synaptic transmission, dopaminergic / cocaine binding / serotonin:sodium:chloride symporter activity / cellular response to cGMP / enteric nervous system development / negative regulation of organ growth / sodium ion binding / neurotransmitter transmembrane transporter activity / serotonin uptake / vasoconstriction / monoamine transmembrane transporter activity / monoamine transport / serotonin binding / brain morphogenesis / syntaxin-1 binding / antiporter activity / negative regulation of neuron differentiation / neurotransmitter transport / male mating behavior / nitric-oxide synthase binding / amino acid transport / membrane depolarization / conditioned place preference / immunoglobulin complex / monoatomic cation channel activity / behavioral response to cocaine / cellular response to retinoic acid / positive regulation of cell cycle / endomembrane system / response to nutrient / sodium ion transmembrane transport / circadian rhythm / platelet aggregation / integrin binding / response to toxic substance / memory / actin filament binding / response to estradiol / presynaptic membrane / adaptive immune response / response to hypoxia / postsynaptic membrane / endosome membrane / neuron projection / membrane raft / response to xenobiotic stimulus / focal adhesion / synapse / positive regulation of gene expression / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
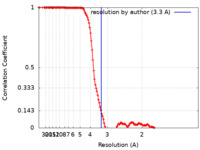
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Coleman JA / Navratna V | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Chemical and structural investigation of the paroxetine-human serotonin transporter complex. Authors: Jonathan A Coleman / Vikas Navratna / Daniele Antermite / Dongxue Yang / James A Bull / Eric Gouaux /   Abstract: Antidepressants target the serotonin transporter (SERT) by inhibiting serotonin reuptake. Structural and biochemical studies aiming to understand binding of small-molecules to conformationally ...Antidepressants target the serotonin transporter (SERT) by inhibiting serotonin reuptake. Structural and biochemical studies aiming to understand binding of small-molecules to conformationally dynamic transporters like SERT often require thermostabilizing mutations and antibodies to stabilize a specific conformation, leading to questions about relationships of these structures to the bonafide conformation and inhibitor binding poses of wild-type transporter. To address these concerns, we determined the structures of ∆N72/∆C13 and ts2-inactive SERT bound to paroxetine analogues using single-particle cryo-EM and x-ray crystallography, respectively. We synthesized enantiopure analogues of paroxetine containing either bromine or iodine instead of fluorine. We exploited the anomalous scattering of bromine and iodine to define the pose of these inhibitors and investigated inhibitor binding to Asn177 mutants of ts2-active SERT. These studies provide mutually consistent insights into how paroxetine and its analogues bind to the central substrate-binding site of SERT, stabilize the outward-open conformation, and inhibit serotonin transport. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21368.map.gz emd_21368.map.gz | 226.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21368-v30.xml emd-21368-v30.xml emd-21368.xml emd-21368.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21368_fsc.xml emd_21368_fsc.xml | 15.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21368.png emd_21368.png | 54 KB | ||
| Filedesc metadata |  emd-21368.cif.gz emd-21368.cif.gz | 6.9 KB | ||
| Others |  emd_21368_additional.map.gz emd_21368_additional.map.gz emd_21368_half_map_1.map.gz emd_21368_half_map_1.map.gz emd_21368_half_map_2.map.gz emd_21368_half_map_2.map.gz | 226.1 MB 57.1 MB 57.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21368 http://ftp.pdbj.org/pub/emdb/structures/EMD-21368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21368 | HTTPS FTP |
-Related structure data
| Related structure data |  6vrhMC  6vrkC  6vrlC  6w2bC  6w2cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10380 (Title: Single particle cryo electron microscopy of the human serotonin transporter bound with paroxetine and in complex with 8B6 Fab EMPIAR-10380 (Title: Single particle cryo electron microscopy of the human serotonin transporter bound with paroxetine and in complex with 8B6 FabData size: 2.7 TB Data #1: Dataset 1 unaligned multi-frame micrographs of SERT-8B6 Fab complex bound to paroxetine [micrographs - multiframe] Data #2: Dataset 2 unaligned multi-frame micrographs of SERT-8B6 Fab complex bound to paroxetine [micrographs - multiframe] Data #3: Aligned micrographs of the SERT-8B6 paroxetine Fab complex bound to paroxetine [micrographs - single frame] Data #4: Particle stack of the SERT-8B6 paroxetine Fab complex bound to paroxetine [picked particles - multiframe - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21368.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21368.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.648 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_21368_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
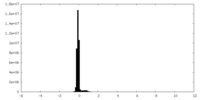
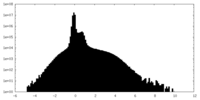
| Density Histograms |
-Half map: halfmap1
| File | emd_21368_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
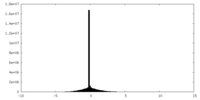
| Density Histograms |
-Half map: halfmap2
| File | emd_21368_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
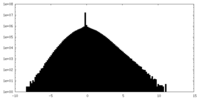
| Density Histograms |
- Sample components
Sample components
+Entire : serotonin transporter 8B6 Fab complex
+Supramolecule #1: serotonin transporter 8B6 Fab complex
+Supramolecule #2: serotonin transporter
+Supramolecule #3: 8B6 antibody
+Macromolecule #1: Sodium-dependent serotonin transporter
+Macromolecule #2: 8B6 antibody, light chain
+Macromolecule #3: 8B6 antibody, heavy chain
+Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #5: Paroxetine
+Macromolecule #6: CHLORIDE ION
+Macromolecule #7: DODECYL-BETA-D-MALTOSIDE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||
| Vitrification | Cryogen name: ETHANE | ||||||||||
| Details | monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)