+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21270 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chloroplast ATP synthase (R1, CF1FO) | |||||||||
 Map data Map data | R1, F1FO | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CF1FO / ATP synthase / PHOTOSYNTHESIS / TRANSLOCASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthetic electron transport in photosystem I / photosynthetic electron transport in photosystem II / chloroplast thylakoid membrane / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism ...photosynthetic electron transport in photosystem I / photosynthetic electron transport in photosystem II / chloroplast thylakoid membrane / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / lipid binding / ATP hydrolysis activity / mitochondrion / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
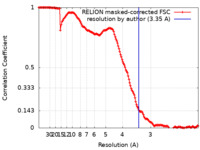
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Yang J-H / Williams D | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Structural basis of redox modulation on chloroplast ATP synthase. Authors: Jay-How Yang / Dewight Williams / Eaazhisai Kandiah / Petra Fromme / Po-Lin Chiu /   Abstract: In higher plants, chloroplast ATP synthase has a unique redox switch on its γ subunit that modulates enzyme activity to limit ATP hydrolysis at night. To understand the molecular details of the ...In higher plants, chloroplast ATP synthase has a unique redox switch on its γ subunit that modulates enzyme activity to limit ATP hydrolysis at night. To understand the molecular details of the redox modulation, we used single-particle cryo-EM to determine the structures of spinach chloroplast ATP synthase in both reduced and oxidized states. The disulfide linkage of the oxidized γ subunit introduces a torsional constraint to stabilize the two β hairpin structures. Once reduced, free cysteines alleviate this constraint, resulting in a concerted motion of the enzyme complex and a smooth transition between rotary states to facilitate the ATP synthesis. We added an uncompetitive inhibitor, tentoxin, in the reduced sample to limit the flexibility of the enzyme and obtained high-resolution details. Our cryo-EM structures provide mechanistic insight into the redox modulation of the energy regulation activity of chloroplast ATP synthase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21270.map.gz emd_21270.map.gz | 18.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21270-v30.xml emd-21270-v30.xml emd-21270.xml emd-21270.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21270_fsc.xml emd_21270_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_21270.png emd_21270.png | 53.3 KB | ||
| Filedesc metadata |  emd-21270.cif.gz emd-21270.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21270 http://ftp.pdbj.org/pub/emdb/structures/EMD-21270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21270 | HTTPS FTP |
-Related structure data
| Related structure data |  6vonMC  6vm1C  6vm4C  6vmbC  6vmdC  6vmgC  6vofC  6vogC  6vohC  6voiC  6vojC  6vokC  6volC  6vomC  6vooC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10475 (Title: Structural basis of redox modulation on chloroplast ATP synthase (reduced form) EMPIAR-10475 (Title: Structural basis of redox modulation on chloroplast ATP synthase (reduced form)Data size: 109.9 Data #1: Reduced state of the chloroplast ATP synthase [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21270.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21270.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | R1, F1FO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.053 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Chloroplast ATP synthase
+Supramolecule #1: Chloroplast ATP synthase
+Macromolecule #1: ATP synthase subunit alpha, chloroplastic
+Macromolecule #2: ATP synthase subunit beta, chloroplastic
+Macromolecule #3: ATP synthase delta chain, chloroplastic
+Macromolecule #4: ATP synthase gamma chain, chloroplastic
+Macromolecule #5: ATP synthase epsilon chain, chloroplastic
+Macromolecule #6: ATP synthase subunit b, chloroplastic
+Macromolecule #7: ATP synthase subunit b', chloroplastic
+Macromolecule #8: ATP synthase subunit a, chloroplastic
+Macromolecule #9: ATP synthase subunit c, chloroplastic
+Macromolecule #10: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #11: TENTOXIN
+Macromolecule #12: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-2/2 4C / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.8000000000000003 µm / Nominal defocus min: -1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)