+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21224 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of Hrd3/Yos9 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | retro-translocation / ERAD / protein degradation / ubiquitination / glycan recognition / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationubiquitin-dependent glycoprotein ERAD pathway / Hrd1p ubiquitin ligase ERAD-M complex / detection of unfolded protein / luminal surveillance complex / Hrd1p ubiquitin ligase complex / Hrd1p ubiquitin ligase ERAD-L complex / negative regulation of protein autoubiquitination / oligosaccharide binding / retrograde protein transport, ER to cytosol / endoplasmic reticulum unfolded protein response ...ubiquitin-dependent glycoprotein ERAD pathway / Hrd1p ubiquitin ligase ERAD-M complex / detection of unfolded protein / luminal surveillance complex / Hrd1p ubiquitin ligase complex / Hrd1p ubiquitin ligase ERAD-L complex / negative regulation of protein autoubiquitination / oligosaccharide binding / retrograde protein transport, ER to cytosol / endoplasmic reticulum unfolded protein response / ERAD pathway / endoplasmic reticulum lumen / endoplasmic reticulum membrane / endoplasmic reticulum / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Wu X / Rapoport TA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Authors: Xudong Wu / Marc Siggel / Sergey Ovchinnikov / Wei Mi / Vladimir Svetlov / Evgeny Nudler / Maofu Liao / Gerhard Hummer / Tom A Rapoport /   Abstract: Misfolded luminal endoplasmic reticulum (ER) proteins undergo ER-associated degradation (ERAD-L): They are retrotranslocated into the cytosol, polyubiquitinated, and degraded by the proteasome. ERAD- ...Misfolded luminal endoplasmic reticulum (ER) proteins undergo ER-associated degradation (ERAD-L): They are retrotranslocated into the cytosol, polyubiquitinated, and degraded by the proteasome. ERAD-L is mediated by the Hrd1 complex (composed of Hrd1, Hrd3, Der1, Usa1, and Yos9), but the mechanism of retrotranslocation remains mysterious. Here, we report a structure of the active Hrd1 complex, as determined by cryo-electron microscopy analysis of two subcomplexes. Hrd3 and Yos9 jointly create a luminal binding site that recognizes glycosylated substrates. Hrd1 and the rhomboid-like Der1 protein form two "half-channels" with cytosolic and luminal cavities, respectively, and lateral gates facing one another in a thinned membrane region. These structures, along with crosslinking and molecular dynamics simulation results, suggest how a polypeptide loop of an ERAD-L substrate moves through the ER membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21224.map.gz emd_21224.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21224-v30.xml emd-21224-v30.xml emd-21224.xml emd-21224.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21224.png emd_21224.png | 104.6 KB | ||
| Filedesc metadata |  emd-21224.cif.gz emd-21224.cif.gz | 6.1 KB | ||
| Others |  emd_21224_additional.map.gz emd_21224_additional.map.gz | 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21224 http://ftp.pdbj.org/pub/emdb/structures/EMD-21224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21224 | HTTPS FTP |
-Related structure data
| Related structure data |  6vk3MC  6vjyC  6vjzC  6vk0C  6vk1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21224.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21224.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_21224_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
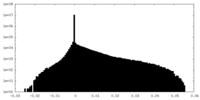
| Density Histograms |
- Sample components
Sample components
-Entire : a complex of Hrd1/Hrd3/Yos9
| Entire | Name: a complex of Hrd1/Hrd3/Yos9 |
|---|---|
| Components |
|
-Supramolecule #1: a complex of Hrd1/Hrd3/Yos9
| Supramolecule | Name: a complex of Hrd1/Hrd3/Yos9 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Hrd3
| Macromolecule | Name: Hrd3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.328023 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MITLLLYLCV ICNAIVLIRA DSIADPWPEA RHLLNTIAKS RDPMKEAAME PNADEFVGFY VPMDYSPRNE EKNYQSIWQN EITDSQRHI YELLVQSSEQ FNNSEATYTL SQIHLWSQYN FPHNMTLAHK YLEKFNDLTH FTNHSAIFDL AVMYATGGCA S GNDQTVIP ...String: MITLLLYLCV ICNAIVLIRA DSIADPWPEA RHLLNTIAKS RDPMKEAAME PNADEFVGFY VPMDYSPRNE EKNYQSIWQN EITDSQRHI YELLVQSSEQ FNNSEATYTL SQIHLWSQYN FPHNMTLAHK YLEKFNDLTH FTNHSAIFDL AVMYATGGCA S GNDQTVIP QDSAKALLYY QRAAQLGNLK AKQVLAYKYY SGFNVPRNFH KSLVLYRDIA EQLRKSYSRD EWDIVFPYWE SY NVRISDF ESGLLGKGLN SVPSSTVRKR TTR(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) DASERRIIRI YYAAL NDYK GTYSQSRNCE RAKNLLELTY KEFQPHVDNL DPLQVFYYVR CLQLLGHMYF TGEGSSKPNI HMAEEILTTS LEISRR AQG PIGRACIDLG LINQYITNNI SQAISYYMKA MKTQANNGIV EFQLSKLATS FPEEKIGDPF NLMETAYLNG FIPAIYE FA VMIESGMNSK SSVENTAYLF KTFVDKNEAI MAPKLRTAFA ALINDRSEVA LWAYSQLAEQ GYETAQVSAA YLMYQLPY E FEDPPRTTDQ RKTLAISYYT RAFKQGNIDA GVVAGDIYFQ MQNYSKAMAL YQGAALKYSI QAIWNLGYMH EHGLGVNRD FHLAKRYYDQ VSEHDHRFYL ASKLSVLKLH LKSWLTWITR EKVNYWKPSS PLNPNEDTQH SKTSWYKQLT KILQRMRHKE DSDKAAEDS HKHRTVVQNG ANHRGDDQEE ASEILGFQME D |
-Macromolecule #2: Protein OS-9 homolog
| Macromolecule | Name: Protein OS-9 homolog / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.311285 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQAKIIYALS AISALIPLGS SLLAPIEDPI VSNKYLISYI DEDDWSDRIL QNQSVMNSGY IVNMGDDLEC FIQNASTQLN DVLEDSNEH SNSEKTALLT KTLNQGVKTI FDKLNERCIF YQAGFWIYEY CPGIEFVQFH GRVNTKTGEI VNRDESLVYR L GKPKANVE ...String: MQAKIIYALS AISALIPLGS SLLAPIEDPI VSNKYLISYI DEDDWSDRIL QNQSVMNSGY IVNMGDDLEC FIQNASTQLN DVLEDSNEH SNSEKTALLT KTLNQGVKTI FDKLNERCIF YQAGFWIYEY CPGIEFVQFH GRVNTKTGEI VNRDESLVYR L GKPKANVE EREFELLYDD VGYYISEIIG SGDICDVTGA ERMVEIQYVC GGSNSGPSTI QWVRETKICV YEAQVTIPEL CN LELLAKN EDQKNASPIL CRMPAKSKIG SNSIDLITKY EPIFLGSGIY FLRPFNTDER DKLMVTDNAM SNWDEITETY YQK FGNAIN KMLSLRLVSL PNGHILQPGD SCVWLAEVVD MKDRFQTTLS LNILNSQRAE IFFNKTFTFN EDNGNFLSYK IGDH GESTE LGQITHSNKA DINTAEIRSD EYLINTDNEL FLRISKEIAE VKELLNEIVS PHEMEVIFEN MRNQPNNDFE LALMN KLKS SLNDDNKVEQ INNARMDDDE STSHTTRDIG EAGSQTTGNT ESEVTNVAAG VFIEHDEL UniProtKB: Protein OS-9 homolog |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 44.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: RANDOM CONICAL TILT |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 99298 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)