+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21140 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a dimer of undecameric human CALHM2 | |||||||||
 Map data Map data | Dimer of undecameric human calcium homeostasis modulator protein 2, sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | taste / assembly / calcium / gap junction / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of microglial cell activation / ATP export / calcium ion import / monoatomic cation channel activity / regulation of synaptic plasticity / positive regulation of apoptotic process / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Syrjanen JL / Chou TH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Structure and assembly of calcium homeostasis modulator proteins. Authors: Johanna L Syrjanen / Kevin Michalski / Tsung-Han Chou / Timothy Grant / Shanlin Rao / Noriko Simorowski / Stephen J Tucker / Nikolaus Grigorieff / Hiro Furukawa /   Abstract: The biological membranes of many cell types contain large-pore channels through which a wide variety of ions and metabolites permeate. Examples include connexin, innexin and pannexin, which form gap ...The biological membranes of many cell types contain large-pore channels through which a wide variety of ions and metabolites permeate. Examples include connexin, innexin and pannexin, which form gap junctions and/or bona fide cell surface channels. The most recently identified large-pore channels are the calcium homeostasis modulators (CALHMs), through which ions and ATP permeate in a voltage-dependent manner to control neuronal excitability, taste signaling and pathologies of depression and Alzheimer's disease. Despite such critical biological roles, the structures and patterns of their oligomeric assembly remain unclear. Here, we reveal the structures of two CALHMs, chicken CALHM1 and human CALHM2, by single-particle cryo-electron microscopy (cryo-EM), which show novel assembly of the four transmembrane helices into channels of octamers and undecamers, respectively. Furthermore, molecular dynamics simulations suggest that lipids can favorably assemble into a bilayer within the larger CALHM2 pore, but not within CALHM1, demonstrating the potential correlation between pore size, lipid accommodation and channel activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21140.map.gz emd_21140.map.gz | 120.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21140-v30.xml emd-21140-v30.xml emd-21140.xml emd-21140.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21140.png emd_21140.png | 156.4 KB | ||
| Filedesc metadata |  emd-21140.cif.gz emd-21140.cif.gz | 5.3 KB | ||
| Others |  emd_21140_additional.map.gz emd_21140_additional.map.gz | 119.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21140 http://ftp.pdbj.org/pub/emdb/structures/EMD-21140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21140 | HTTPS FTP |
-Related structure data
| Related structure data |  6vaiMC  6vakC  6valC  6vamC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10487 (Title: Human CALHM2 gap junction in nanodisc, 1 mM CaCl2 and 1 mM ATP EMPIAR-10487 (Title: Human CALHM2 gap junction in nanodisc, 1 mM CaCl2 and 1 mM ATPData size: 216.5 Data #1: Unaligned movies (.tif) for human CALHM2 gap junction in nanodisc at 1 mM CaCl2 and 1 mM ATP [micrographs - multiframe])  EMPIAR-10488 (Title: Human CALHM2 in nanodisc, 1 mM EDTA / Data size: 604.3 EMPIAR-10488 (Title: Human CALHM2 in nanodisc, 1 mM EDTA / Data size: 604.3 Data #1: Unaligned movies (.tif) for human CALHM2 in nanodisc at 1 mM EDTA [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21140.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21140.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer of undecameric human calcium homeostasis modulator protein 2, sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
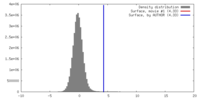
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_21140_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
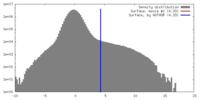
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of calcium homeostasis modulator protein 2 (CALHM2) undecam...
| Entire | Name: Dimer of calcium homeostasis modulator protein 2 (CALHM2) undecamers (22-mer; gap junction like) |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of calcium homeostasis modulator protein 2 (CALHM2) undecam...
| Supramolecule | Name: Dimer of calcium homeostasis modulator protein 2 (CALHM2) undecamers (22-mer; gap junction like) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcium homeostasis modulator protein 2
| Macromolecule | Name: Calcium homeostasis modulator protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 22 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.514957 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAWSHPQFE KGGGSGGGSG GSAWSHPQFE KGGTGAALIA ENFRFLSLFF KSKDVMIFNG LVALGTVGSQ ELFSVVAFHC PCSPARNYL YGLAAIGVPA LVLFIIGIIL NNHTWNLVAE CQHRRTKNCS AAPTFLLLSS ILGRAAVAPV TWSVISLLRG E AYVCALSE ...String: MSAWSHPQFE KGGGSGGGSG GSAWSHPQFE KGGTGAALIA ENFRFLSLFF KSKDVMIFNG LVALGTVGSQ ELFSVVAFHC PCSPARNYL YGLAAIGVPA LVLFIIGIIL NNHTWNLVAE CQHRRTKNCS AAPTFLLLSS ILGRAAVAPV TWSVISLLRG E AYVCALSE FVDPSSLTAR EEHFPSAHAT EILARFPCKE NPDNLSDFRE EVSRRLRYES QLFGWLLIGV VAILVFLTKC LK HYCSPLS YRQEAYWAQY RANEDQLFQR TAEVHSRVLA ANNVRRFFGF VALNKDDEEL IANFPVEGTQ PRPQWNAITG VYL YRENQG LPLYSRLHKW AQGLAGNGAA PDNVEMALLP S UniProtKB: Calcium homeostasis modulator protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 288.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4 sec before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: undecameric human CALHM2 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D11 (2x11 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 3.68 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 52737 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)