+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21136 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse retromer (VPS26/VPS35/VPS29) heterotrimer | |||||||||
 Map data Map data | Mouse retromer (VPS26/VPS35/VPS29) heterotrimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | retromer / membrane trafficking / endosomal trafficking / membrane coat complexes / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationWNT ligand biogenesis and trafficking / neurotransmitter receptor transport, endosome to plasma membrane / negative regulation of protein localization / mitochondrion-derived vesicle / regulation of dendritic spine maintenance / negative regulation of protein homooligomerization / tubular endosome / regulation of terminal button organization / positive regulation of Wnt protein secretion / retromer, cargo-selective complex ...WNT ligand biogenesis and trafficking / neurotransmitter receptor transport, endosome to plasma membrane / negative regulation of protein localization / mitochondrion-derived vesicle / regulation of dendritic spine maintenance / negative regulation of protein homooligomerization / tubular endosome / regulation of terminal button organization / positive regulation of Wnt protein secretion / retromer, cargo-selective complex / vacuolar protein processing / mitochondrion to lysosome vesicle-mediated transport / protein localization to organelle / Golgi to vacuole transport / negative regulation of lysosomal protein catabolic process / positive regulation of locomotion involved in locomotory behavior / negative regulation of late endosome to lysosome transport / positive regulation of dopamine receptor signaling pathway / positive regulation of dopamine biosynthetic process / neurotransmitter receptor transport, endosome to postsynaptic membrane / protein localization to endosome / vesicle-mediated transport in synapse / voluntary musculoskeletal movement / retromer complex / mitochondrial fragmentation involved in apoptotic process / transcytosis / dopaminergic synapse / regulation of synapse maturation / endocytic recycling / retrograde transport, endosome to Golgi / positive regulation of protein localization to cell periphery / lysosome organization / positive regulation of mitochondrial fission / regulation of postsynapse assembly / D1 dopamine receptor binding / intracellular protein transport / modulation of chemical synaptic transmission / negative regulation of inflammatory response / protein destabilization / positive regulation of protein catabolic process / late endosome / positive regulation of canonical Wnt signaling pathway / protein transport / presynapse / vesicle / early endosome / lysosome / endosome / endosome membrane / neuron projection / postsynapse / postsynaptic density / negative regulation of gene expression / neuronal cell body / synapse / positive regulation of gene expression / perinuclear region of cytoplasm / glutamatergic synapse / mitochondrion / metal ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
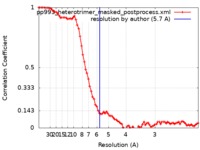
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | |||||||||
 Authors Authors | Kendall AK / Jackson LP | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes. Authors: Amy K Kendall / Boyang Xie / Peng Xu / Jue Wang / Rodger Burcham / Meredith N Frazier / Elad Binshtein / Hui Wei / Todd R Graham / Terunaga Nakagawa / Lauren P Jackson /  Abstract: Metazoan retromer (VPS26/VPS35/VPS29) associates with sorting nexins on endosomal tubules to sort proteins to the trans-Golgi network or plasma membrane. Mechanisms of metazoan retromer assembly ...Metazoan retromer (VPS26/VPS35/VPS29) associates with sorting nexins on endosomal tubules to sort proteins to the trans-Golgi network or plasma membrane. Mechanisms of metazoan retromer assembly remain undefined. We combine single-particle cryoelectron microscopy with biophysical methods to uncover multiple oligomer structures. 2D class averages reveal mammalian heterotrimers; dimers of trimers; tetramers of trimers; and flat chains. These species are further supported by biophysical solution studies. We provide reconstructions of all species, including key sub-structures (∼5 Å resolution). Local resolution variation suggests that heterotrimers and dimers adopt multiple conformations. Our structures identify a flexible, highly conserved electrostatic dimeric interface formed by VPS35 subunits. We generate structure-based mutants to disrupt this interface in vitro. Equivalent mutations in yeast demonstrate a mild cargo-sorting defect. Our data suggest the metazoan retromer is an adaptable and plastic scaffold that accommodates interactions with different sorting nexins to sort multiple cargoes from endosomes their final destinations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21136.map.gz emd_21136.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21136-v30.xml emd-21136-v30.xml emd-21136.xml emd-21136.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21136_fsc.xml emd_21136_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_21136.png emd_21136.png | 140.2 KB | ||
| Filedesc metadata |  emd-21136.cif.gz emd-21136.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21136 http://ftp.pdbj.org/pub/emdb/structures/EMD-21136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21136 | HTTPS FTP |
-Related structure data
| Related structure data |  6vacMC  6vabC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21136.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21136.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mouse retromer (VPS26/VPS35/VPS29) heterotrimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.096 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mouse retromer (VPS26/VPS35/VPS29) tetramer of heterotrimers
| Entire | Name: Mouse retromer (VPS26/VPS35/VPS29) tetramer of heterotrimers |
|---|---|
| Components |
|
-Supramolecule #1: Mouse retromer (VPS26/VPS35/VPS29) tetramer of heterotrimers
| Supramolecule | Name: Mouse retromer (VPS26/VPS35/VPS29) tetramer of heterotrimers type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Vacuolar protein sorting-associated protein 35
| Macromolecule | Name: Vacuolar protein sorting-associated protein 35 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 91.821727 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPTTQQSPQD EQEKLLDEAI QAVKVQSFQM KRCLDKNKLM DALKHASNML GELRTSMLSP KSYYELYMAI SDELHYLEVY LTDEFAKGR KVADLYELVQ YAGNIIPRLY LLITVGVVYV KSFPQSRKDI LKDLVEMCRG VQHPLRGLFL RNYLLQCTRN I LPDEGEPT ...String: MPTTQQSPQD EQEKLLDEAI QAVKVQSFQM KRCLDKNKLM DALKHASNML GELRTSMLSP KSYYELYMAI SDELHYLEVY LTDEFAKGR KVADLYELVQ YAGNIIPRLY LLITVGVVYV KSFPQSRKDI LKDLVEMCRG VQHPLRGLFL RNYLLQCTRN I LPDEGEPT DEETTGDISD SMDFVLLNFA EMNKLWVRMQ HQGHSRDREK RERERQELRI LVGTNLVRLS QLEGVNVERY KQ IVLTGIL EQVVNCRDAL AQEYLMECII QVFPDEFHLQ TLNPFLRACA ELHQNVNVKN IIIALIDRLA LFAHREDGPG IPA EIKLFD IFSQQVATVI QSRQDMPSED VVSLQVSLIN LAMKCYPDRV DYVDKVLETT VEIFNKLNLE HIATSSAVSK ELTR LLKIP VDTYNNILTV LKLKHFHPLF EYFDYESRKS MSCYVLSNVL DYNTEIVSQD QVDSIMNLVS TLIQDQPDQP VEDPD PEDF ADEQSLVGRF IHLLRSDDPD QQYLILNTAR KHFGAGGNQR IRFTLPPLVF AAYQLAFRYK ENSQMDDKWE KKCQKI FSF AHQTISALIK AELAELPLRL FLQGALAAGE IGFENHETVA YEFMSQAFSL YEDEISDSKA QLAAITLIIG TFERMKC FS EENHEPLRTQ CALAASKLLK KPDQGRAVST CAHLFWSGRN TDKNGEELHG GKRVMECLKK ALKIANQCMD PSLQVQLF I EILNRYIYFY EKENDAVTIQ VLNQLIQKIR EDLPNLESSE ETEQINKHFH NTLEHLRSRR ESPESEGPIY EGLIL UniProtKB: Vacuolar protein sorting-associated protein 35 |
-Macromolecule #2: Vacuolar protein sorting-associated protein 26A
| Macromolecule | Name: Vacuolar protein sorting-associated protein 26A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.167789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFLGGFFGP ICEIDVALND GETRKMAEMK TEDGKVEKHY LFYDGESVSG KVNLAFKQPG KRLEHQGIRI EFVGQIELFN DKSNTHEFV NLVKELALPG ELTQSRSYDF EFMQVEKPYE SYIGANVRLR YFLKVTIVRR LTDLVKEYDL IVHQLATYPD V NNSIKMEV ...String: MSFLGGFFGP ICEIDVALND GETRKMAEMK TEDGKVEKHY LFYDGESVSG KVNLAFKQPG KRLEHQGIRI EFVGQIELFN DKSNTHEFV NLVKELALPG ELTQSRSYDF EFMQVEKPYE SYIGANVRLR YFLKVTIVRR LTDLVKEYDL IVHQLATYPD V NNSIKMEV GIEDCLHIEF EYNKSKYHLK DVIVGKIYFL LVRIKIQHME LQLIKKEITG IGPSTTTETE TIAKYEIMDG AP VKGESIP IRLFLAGYDP TPTMRDVNKK FSVRYFLNLV LVDEEDRRYF KQQEIILWRK APEKLRKQRT NFHQRFESPD SQA SAEQPE M UniProtKB: Vacuolar protein sorting-associated protein 26A |
-Macromolecule #3: Vacuolar protein sorting-associated protein 29
| Macromolecule | Name: Vacuolar protein sorting-associated protein 29 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.521668 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLVLVLGDLH IPHRCNSLPA KFKKLLVPGK IQHILCTGNL CTKESYDYLK TLAGDVHIVR GDFDENLNYP EQKVVTVGQF KIGLIHGHQ VIPWGDMASL ALLQRQFDVD ILISGHTHKF EAFEHENKFY INPGSATGAY NALETNIIPS FVLMDIQAST V VTYVYQLI GDDVKVERIE YKKS UniProtKB: Vacuolar protein sorting-associated protein 29 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 71.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)