[English] 日本語
 Yorodumi
Yorodumi- EMDB-21044: Cryo-EM structure of the integrin AlphaIIbBeta3-Abciximab complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21044 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
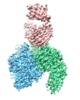
| Title | Cryo-EM structure of the integrin AlphaIIbBeta3-Abciximab complex | ||||||||||||
 Map data Map data | integrin AlphaIIbBeta3-Abciximab complex | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Integrin / Abciximab / BLOOD CLOTTING / Cell Adhesion-Immune System complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of serotonin uptake / positive regulation of adenylate cyclase-inhibiting opioid receptor signaling pathway / tube development / alpha9-beta1 integrin-ADAM8 complex / regulation of trophoblast cell migration / integrin alphaIIb-beta3 complex / regulation of postsynaptic neurotransmitter receptor diffusion trapping / alphav-beta3 integrin-vitronectin complex / maintenance of postsynaptic specialization structure / regulation of extracellular matrix organization ...regulation of serotonin uptake / positive regulation of adenylate cyclase-inhibiting opioid receptor signaling pathway / tube development / alpha9-beta1 integrin-ADAM8 complex / regulation of trophoblast cell migration / integrin alphaIIb-beta3 complex / regulation of postsynaptic neurotransmitter receptor diffusion trapping / alphav-beta3 integrin-vitronectin complex / maintenance of postsynaptic specialization structure / regulation of extracellular matrix organization / positive regulation of glomerular mesangial cell proliferation / platelet alpha granule membrane / integrin alphav-beta3 complex / negative regulation of lipoprotein metabolic process / alphav-beta3 integrin-PKCalpha complex / fibrinogen binding / alphav-beta3 integrin-HMGB1 complex / vascular endothelial growth factor receptor 2 binding / negative regulation of lipid transport / positive regulation of vascular endothelial growth factor signaling pathway / Elastic fibre formation / mesodermal cell differentiation / cell-substrate junction assembly / alphav-beta3 integrin-IGF-1-IGF1R complex / positive regulation of bone resorption / platelet-derived growth factor receptor binding / glycinergic synapse / regulation of release of sequestered calcium ion into cytosol / filopodium membrane / extracellular matrix binding / wound healing, spreading of epidermal cells / positive regulation of vascular endothelial growth factor receptor signaling pathway / apolipoprotein A-I-mediated signaling pathway / positive regulation of cell adhesion mediated by integrin / positive regulation of leukocyte migration / negative regulation of low-density lipoprotein particle clearance / regulation of bone resorption / angiogenesis involved in wound healing / apoptotic cell clearance / positive regulation of fibroblast migration / integrin complex / positive regulation of smooth muscle cell migration / heterotypic cell-cell adhesion / smooth muscle cell migration / Molecules associated with elastic fibres / cell adhesion mediated by integrin / negative chemotaxis / positive regulation of cell-matrix adhesion / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / Syndecan interactions / p130Cas linkage to MAPK signaling for integrins / cellular response to insulin-like growth factor stimulus / protein disulfide isomerase activity / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of osteoblast proliferation / microvillus membrane / cell-substrate adhesion / platelet-derived growth factor receptor signaling pathway / PECAM1 interactions / GRB2:SOS provides linkage to MAPK signaling for Integrins / TGF-beta receptor signaling activates SMADs / fibronectin binding / lamellipodium membrane / blood coagulation, fibrin clot formation / negative regulation of macrophage derived foam cell differentiation / negative regulation of lipid storage / ECM proteoglycans / Integrin cell surface interactions / negative regulation of endothelial cell apoptotic process / positive regulation of T cell migration / coreceptor activity / cellular response to platelet-derived growth factor stimulus / positive regulation of endothelial cell proliferation / Integrin signaling / cell adhesion molecule binding / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of endothelial cell migration / positive regulation of smooth muscle cell proliferation / substrate adhesion-dependent cell spreading / embryo implantation / protein kinase C binding / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / cell-matrix adhesion / response to activity / integrin-mediated signaling pathway / Signal transduction by L1 / regulation of actin cytoskeleton organization / wound healing / cellular response to mechanical stimulus / cell-cell adhesion / Signaling by high-kinase activity BRAF mutants / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / MAP2K and MAPK activation / platelet activation / VEGFA-VEGFR2 Pathway / platelet aggregation / integrin binding / cellular response to xenobiotic stimulus / ruffle membrane / positive regulation of fibroblast proliferation Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Nesic D / Zhang Y | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Arterioscler Thromb Vasc Biol / Year: 2020 Journal: Arterioscler Thromb Vasc Biol / Year: 2020Title: Cryo-Electron Microscopy Structure of the αIIbβ3-Abciximab Complex. Authors: Dragana Nešić / Yixiao Zhang / Aleksandar Spasic / Jihong Li / Davide Provasi / Marta Filizola / Thomas Walz / Barry S Coller Abstract: OBJECTIVE: The αIIbβ3 antagonist antiplatelet drug abciximab is the chimeric antigen-binding fragment comprising the variable regions of murine monoclonal antibody 7E3 and the constant domains of ...OBJECTIVE: The αIIbβ3 antagonist antiplatelet drug abciximab is the chimeric antigen-binding fragment comprising the variable regions of murine monoclonal antibody 7E3 and the constant domains of human IgG1 and light chain κ. Previous mutagenesis studies suggested that abciximab binds to the β3 C177-C184 specificity-determining loop (SDL) and Trp129 on the adjacent β1-α1 helix. These studies could not, however, assess whether 7E3 or abciximab prevents fibrinogen binding by steric interference, disruption of either the αIIbβ3-binding pocket for fibrinogen or the β3 SDL (which is not part of the binding pocket but affects fibrinogen binding), or some combination of these effects. To address this gap, we used cryo-electron microscopy to determine the structure of the αIIbβ3-abciximab complex at 2.8 Å resolution. Approach and Results: The interacting surface of abciximab is comprised of residues from all 3 complementarity-determining regions of both the light and heavy chains, with high representation of aromatic residues. Binding is primarily to the β3 SDL and neighboring residues, the β1-α1 helix, and β3 residues Ser211, Val212 and Met335. Unexpectedly, the structure also indicated several interactions with αIIb. As judged by the cryo-electron microscopy model, molecular-dynamics simulations, and mutagenesis, the binding of abciximab does not appear to rely on the interaction with the αIIb residues and does not result in disruption of the fibrinogen-binding pocket; it does, however, compress and reduce the flexibility of the SDL. CONCLUSIONS: We deduce that abciximab prevents ligand binding by steric interference, with a potential contribution via displacement of the SDL and limitation of the flexibility of the SDL residues. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21044.map.gz emd_21044.map.gz | 57.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21044-v30.xml emd-21044-v30.xml emd-21044.xml emd-21044.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21044.png emd_21044.png | 137.7 KB | ||
| Filedesc metadata |  emd-21044.cif.gz emd-21044.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21044 http://ftp.pdbj.org/pub/emdb/structures/EMD-21044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21044 | HTTPS FTP |
-Related structure data
| Related structure data |  6v4pMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21044.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21044.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | integrin AlphaIIbBeta3-Abciximab complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Integrin AlphaIIbBeta3-Abciximab Complex
| Entire | Name: Integrin AlphaIIbBeta3-Abciximab Complex |
|---|---|
| Components |
|
-Supramolecule #1: Integrin AlphaIIbBeta3-Abciximab Complex
| Supramolecule | Name: Integrin AlphaIIbBeta3-Abciximab Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Integrin alpha-IIb
| Macromolecule | Name: Integrin alpha-IIb / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 104.894438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARALCPLQA LWLLEWVLLL LGPCAAPPAW ALNLDPVQLT FYAGPNGSQF GFSLDFHKDS HGRVAIVVGA PRTLGPSQEE TGGVFLCPW RAEGGQCPSL LFDLRDETRN VGSQTLQTFK ARQGLGASVV SWSDVIVACA PWQHWNVLEK TEEAEKTPVG S CFLAQPES ...String: MARALCPLQA LWLLEWVLLL LGPCAAPPAW ALNLDPVQLT FYAGPNGSQF GFSLDFHKDS HGRVAIVVGA PRTLGPSQEE TGGVFLCPW RAEGGQCPSL LFDLRDETRN VGSQTLQTFK ARQGLGASVV SWSDVIVACA PWQHWNVLEK TEEAEKTPVG S CFLAQPES GRRAEYSPCR GNTLSRIYVE NDFSWDKRYC EAGFSSVVTQ AGELVLGAPG GYYFLGLLAQ APVADIFSSY RP GILLWHV SSQSLSFDSS NPEYFDGYWG YSVAVGEFDG DLNTTEYVVG APTWSWTLGA VEILDSYYQR LHRLRGEQMA SYF GHSVAV TDVNGDGRHD LLVGAPLYME SRADRKLAEV GRVYLFLQPR GPHALGAPSL LLTGTQLYGR FGSAIAPLGD LDRD GYNDI AVAAPYGGPS GRGQVLVFLG QSEGLRSRPS QVLDSPFPTG SAFGFSLRGA VDIDDNGYPD LIVGAYGANQ VAVYR AQPV VKASVQLLVQ DSLNPAVKSC VLPQTKTPVS CFNIQMCVGA TGHNIPQKLS LNAELQLDRQ KPRQGRRVLL LGSQQA GTT LNLDLGGKHS PICHTTMAFL RDEADFRDKL SPIVLSLNVS LPPTEAGMAP AVVLHGDTHV QEQTRIVLDC GEDDVCV PQ LQLTASVTGS PLLVGADNVL ELQMDAANEG EGAYEAELAV HLPQGAHYMR ALSNVEGFER LICNQKKENE TRVVLCEL G NPMKKNAQIG IAMLVSVGNL EEAGESVSFQ LQIRSKNSQN PNSKIVLLDV PVRAEAQVEL RGNSFPASLV VAAEEGERE QNSLDSWGPK VEHTYELHNN GPGTVNGLHL SIHLPGQSQP SDLLYILDIQ PQGGLQCFPQ PPVNPLKVDW GLPIPSPSPI HPAHHKRDR RQIFLPEPEQ PSRLQDPVLV SCDSAPCTVV QCDLQEMARG QRAMVTVLAF LWLPSLYQRP LDQFVLQSHA W FNV UniProtKB: Integrin alpha-IIb |
-Macromolecule #2: Integrin beta-3
| Macromolecule | Name: Integrin beta-3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.974828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRARPRPRPL WATVLALGAL AGVGVGGPNI CTTRGVSSCQ QCLAVSPMCA WCSDEALPLG SPRCDLKENL LKDNCAPESI EFPVSEARV LEDRPLSDKG SGDSSQVTQV SPQRIALRLR PDDSKNFSIQ VRQVEDYPVD IYYLMDLSYS MKDDLWSIQN L GTKLATQM ...String: MRARPRPRPL WATVLALGAL AGVGVGGPNI CTTRGVSSCQ QCLAVSPMCA WCSDEALPLG SPRCDLKENL LKDNCAPESI EFPVSEARV LEDRPLSDKG SGDSSQVTQV SPQRIALRLR PDDSKNFSIQ VRQVEDYPVD IYYLMDLSYS MKDDLWSIQN L GTKLATQM RKLTSNLRIG FGAFVDKPVS PYMYISPPEA LENPCYDMKT TCLPMFGYKH VLTLTDQVTR FNEEVKKQSV SR NRDAPEG GFDAIMQATV CDEKIGWRND ASHLLVFTTD AKTHIALDGR LAGIVQPNDG QCHVGSDNHY SASTTMDYPS LGL MTEKLS QKNINLIFAV TENVVNLYQN YSELIPGTTV GVLSMDSSNV LQLIVDAYGK IRSKVELEVR DLPEELSLSF NATC LNNEV IPGLKSCMGL KIGDTVSFSI EAKVRGCPQE KEKSFTIKPV GFKDSLIVQV TFDCDCACQA QAEPNSHRCN NGNGT FECG VCRCGPGWLG SQCECSEEDY RPSQQDECSP REGQPVCSQR GECLCGQCVC HSSDFGKITG KYCECDDFSC VRYKGE MCS GHGQCSCGDC LCDSDWTGYY CNCTTRTDTC MSSNGLLCSG RGKCECGSCV CIQPGSYGDT CEKCPTCPDA CTFKKEC VE CKKFDRGALH DENTCNRYCR DEIESVKELK DTGKDAVNCT YKNEDDCV UniProtKB: Integrin beta-3 |
-Macromolecule #3: Abciximab, heavy chain
| Macromolecule | Name: Abciximab, heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 24.161119 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLQQSGTV LARPGASVKM SCEASGYTFT NYWMHWVKQR PGQGLEWIGA IYPGNSDTSY IQKFKGKAKL TAVTSTTSVY MELSSLTNE DSAVYYCTLY DGYYVFAYWG QGTLVTVSAA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV ...String: EVQLQQSGTV LARPGASVKM SCEASGYTFT NYWMHWVKQR PGQGLEWIGA IYPGNSDTSY IQKFKGKAKL TAVTSTTSVY MELSSLTNE DSAVYYCTLY DGYYVFAYWG QGTLVTVSAA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP KSCDKTH |
-Macromolecule #4: Abciximab, light chain
| Macromolecule | Name: Abciximab, light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.501986 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPVT LSVTPGDSVS LSCRASRDIS NNLHWFQQTS HESPRLLIKY ASQSMSGIPS RFSGSGSGTD FTLSINSVET EDFGMYFCQ QTNSWPYTFG GGTKLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: EIVLTQSPVT LSVTPGDSVS LSCRASRDIS NNLHWFQQTS HESPRLLIKY ASQSMSGIPS RFSGSGSGTD FTLSINSVET EDFGMYFCQ QTNSWPYTFG GGTKLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















