+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20949 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cardiac sodium channel with flecainide | |||||||||
 Map data Map data | CryoEM map for a drug binding structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cardiac sodium channel complex with flecainide / METAL TRANSPORT / ion channel | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated sodium channel activity involved in AV node cell action potential / voltage-gated sodium channel activity involved in bundle of His cell action potential / voltage-gated sodium channel activity involved in Purkinje myocyte action potential / voltage-gated sodium channel activity involved in SA node cell action potential / bundle of His cell action potential / regulation of ventricular cardiac muscle cell membrane depolarization / AV node cell action potential / SA node cell action potential / AV node cell to bundle of His cell communication / membrane depolarization during SA node cell action potential ...voltage-gated sodium channel activity involved in AV node cell action potential / voltage-gated sodium channel activity involved in bundle of His cell action potential / voltage-gated sodium channel activity involved in Purkinje myocyte action potential / voltage-gated sodium channel activity involved in SA node cell action potential / bundle of His cell action potential / regulation of ventricular cardiac muscle cell membrane depolarization / AV node cell action potential / SA node cell action potential / AV node cell to bundle of His cell communication / membrane depolarization during SA node cell action potential / response to denervation involved in regulation of muscle adaptation / sodium channel complex / membrane depolarization during atrial cardiac muscle cell action potential / cardiac ventricle development / regulation of atrial cardiac muscle cell membrane repolarization / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / brainstem development / membrane depolarization during AV node cell action potential / membrane depolarization during bundle of His cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / positive regulation of action potential / atrial cardiac muscle cell action potential / membrane depolarization during Purkinje myocyte cell action potential / telencephalon development / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / ventricular cardiac muscle cell action potential / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / voltage-gated sodium channel complex / sodium ion import across plasma membrane / regulation of cardiac muscle cell contraction / ankyrin binding / voltage-gated sodium channel activity / sodium ion transport / nitric-oxide synthase binding / fibroblast growth factor binding / odontogenesis of dentin-containing tooth / regulation of heart rate by cardiac conduction / intercalated disc / lateral plasma membrane / membrane depolarization / positive regulation of heart rate / neuronal action potential / cardiac muscle contraction / T-tubule / sodium ion transmembrane transport / regulation of heart rate / cellular response to calcium ion / bioluminescence / cerebellum development / positive regulation of epithelial cell proliferation / generation of precursor metabolites and energy / sarcolemma / caveola / Z disc / scaffold protein binding / transmembrane transporter binding / calmodulin binding / protein domain specific binding / axon / ubiquitin protein ligase binding / protein kinase binding / perinuclear region of cytoplasm / enzyme binding / cell surface / endoplasmic reticulum / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
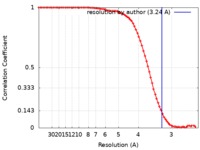
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Jiang D / Shi H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Structure of the Cardiac Sodium Channel. Authors: Daohua Jiang / Hui Shi / Lige Tonggu / Tamer M Gamal El-Din / Michael J Lenaeus / Yan Zhao / Craig Yoshioka / Ning Zheng / William A Catterall /  Abstract: Voltage-gated sodium channel Na1.5 generates cardiac action potentials and initiates the heartbeat. Here, we report structures of Na1.5 at 3.2-3.5 Å resolution. Na1.5 is distinguished from other ...Voltage-gated sodium channel Na1.5 generates cardiac action potentials and initiates the heartbeat. Here, we report structures of Na1.5 at 3.2-3.5 Å resolution. Na1.5 is distinguished from other sodium channels by a unique glycosyl moiety and loss of disulfide-bonding capability at the Naβ subunit-interaction sites. The antiarrhythmic drug flecainide specifically targets the central cavity of the pore. The voltage sensors are partially activated, and the fast-inactivation gate is partially closed. Activation of the voltage sensor of Domain III allows binding of the isoleucine-phenylalanine-methionine (IFM) motif to the inactivation-gate receptor. Asp and Ala, in the selectivity motif DEKA, line the walls of the ion-selectivity filter, whereas Glu and Lys are in positions to accept and release Na ions via a charge-delocalization network. Arrhythmia mutation sites undergo large translocations during gating, providing a potential mechanism for pathogenic effects. Our results provide detailed insights into Na1.5 structure, pharmacology, activation, inactivation, ion selectivity, and arrhythmias. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20949.map.gz emd_20949.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20949-v30.xml emd-20949-v30.xml emd-20949.xml emd-20949.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20949_fsc.xml emd_20949_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20949.png emd_20949.png | 63.2 KB | ||
| Filedesc metadata |  emd-20949.cif.gz emd-20949.cif.gz | 7.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20949 http://ftp.pdbj.org/pub/emdb/structures/EMD-20949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20949 | HTTPS FTP |
-Validation report
| Summary document |  emd_20949_validation.pdf.gz emd_20949_validation.pdf.gz | 699.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20949_full_validation.pdf.gz emd_20949_full_validation.pdf.gz | 699.3 KB | Display | |
| Data in XML |  emd_20949_validation.xml.gz emd_20949_validation.xml.gz | 10.6 KB | Display | |
| Data in CIF |  emd_20949_validation.cif.gz emd_20949_validation.cif.gz | 13.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20949 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20949 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20949 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20949 | HTTPS FTP |
-Related structure data
| Related structure data |  6uz0MC  6uz3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20949.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20949.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map for a drug binding structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.056 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : cardiac sodium channel complex with flecainide
| Entire | Name: cardiac sodium channel complex with flecainide |
|---|---|
| Components |
|
-Supramolecule #1: cardiac sodium channel complex with flecainide
| Supramolecule | Name: cardiac sodium channel complex with flecainide / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Sodium channel protein type 5 subunit alpha,Green fluorescent protein
| Macromolecule | Name: Sodium channel protein type 5 subunit alpha,Green fluorescent protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 209.037781 KDa |
| Recombinant expression | Organism:  Mammalian 1 orthobornavirus Mammalian 1 orthobornavirus |
| Sequence | String: MANLLLPRGT SSFRRFTRES LAAIEKRMAE KQARGGSATS QESREGLQEE EAPRPQLDLQ ASKKLPDLYG NPPRELIGEP LEDLDPFYS TQKTFIVLNK GKTIFRFSAT NALYVLSPFH PVRRAAVKIL VHSLFSMLIM CTILTNCVFM AQHDPPPWTK Y VEYTFTAI ...String: MANLLLPRGT SSFRRFTRES LAAIEKRMAE KQARGGSATS QESREGLQEE EAPRPQLDLQ ASKKLPDLYG NPPRELIGEP LEDLDPFYS TQKTFIVLNK GKTIFRFSAT NALYVLSPFH PVRRAAVKIL VHSLFSMLIM CTILTNCVFM AQHDPPPWTK Y VEYTFTAI YTFESLVKIL ARGFCLHAFT FLRDPWNWLD FSVIVMAYTT EFVDLGNVSA LRTFRVLRAL KTISVISGLK TI VGALIQS VKKLADVMVL TVFCLSVFAL IGLQLFMGNL RHKCVRNFTE LNGTNGSVEA DGLVWNSLDV YLNDPANYLL KNG TTDVLL CGNSSDAGTC PEGYRCLKAG ENPDHGYTSF DSFAWAFLAL FRLMTQDCWE RLYQQTLRSA GKIYMIFFML VIFL GSFYL VNLILAVVAM AYEEQNQATI AETEEKEKRF QEAMEMLKKE HEALTIRGVD TVSRSSARQR ALSAVSVLTS ALEEL EESH RKCPPCWNRF AQHYLIWECC PLWMSIKQKV KFVVMDPFAD LTITMCIVLN TLFMALEHYN MTAEFEEMLQ VGNLVF TGI FTAEMTFKII ALDPYYYFQQ GWNIFDSIIV ILSLMELGLS RMGNLSVLRS FRLLRVFKLA KSWPTLNTLI KIIGNSV GA LGNLTLVLAI IVFIFAVVGM QLFGKNYSEL RHRISDSGLL PRWHMMDFFH AFLIIFRILC GEWIETMWDC MEVSGQSL C LLVFLLVMVI GNLVVLNLFL ALLLSSFSAD NLTAPDEDGE MNNLQLALAR IQRGLRFVKR TTWDFCCGIL RRRPKKPAA LATHSQLPSC ITAPRSPPPP EVEKVPPARK ETRFEEDKRP GQGTPGDSEP VCVPIAVAES DTEDQEEDEE NGKVWWRLRK TCYRIVEHS WFETFIIFMI LLSSGALAFE DIYLEERKTI KVLLEYADKM FTYVFVLEML LKWVAYGFKK YFTNAWCWLD F LIVDVSLV SLVANTLGFA EMGPIKSLRT LRALRPLRAL SRFEGMRVVV NALVGAIPSI MNVLLVCLIF WLIFSIMGVN LF AGKFGRC INQTEGDLPL NYTIVNNKSE CESFNVTGEL YWTKVKVNFD NVGAGYLALL QVATFKGWMD IMYAAVDSRG YEE QPQWED NLYMYIYFVV FIIFGSFFTL NLFIGVIIDN FNQQKKKLGG QDIFMTEEQK KYYNAMKKLG SKKPQKPIPR PLNK YQGFI FDIVTKQAFD VTIMFLICLN MVTMMVETDD QSPEKVNILA KINLLFVAIF TGECIVKMAA LRHYYFTNSW NIFDF VVVI LSIVGTVLSD IIQKYFFSPT LFRVIRLARI GRILRLIRGA KGIRTLLFAL MMSLPALFNI GLLLFLVMFI YSIFGM ANF AYVKWEAGID DMFNFQTFAN SMLCLFQITT SAGWDGLLSP ILNTGPPYCD PNLPNSNGSR GNCGSPAVGI LFFTTYI II SFLIVVNMYI AIILENFSVA TEESTEPLSE DDFDMFYEIW EKFDPEATQF IEYLALSDFA DALSEPLRIA KPNQISLI N MDLPMVSGDR IHCMDILFAF TKRVLGESGE MDALKIQMEE KFMAANPSKI SYEPITTTLE VLFQGPGSMV SKGEELFTG VVPILVELDG DVNGHKFSVS GEGEGDATYG KLTLKFICTT GKLPVPWPTL VTTLTYGVQC FSRYPDHMKQ HDFFKSAMPE GYVQERTIF FKDDGNYKTR AEVKFEGDTL VNRIELKGID FKEDGNILGH KLEYNYNSHN VYIMADKQKN GIKVNFKIRH N IEDGSVQL ADHYQQNTPI GDGPVLLPDN HYLSTQSALS KDPNEKRDHM VLLEFVTAAG ITLGMDELYK GSDYKDDDDK UniProtKB: Sodium channel protein type 5 subunit alpha, Green fluorescent protein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 5 / Number of copies: 5 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Macromolecule #6: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 6 / Number of copies: 11 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Macromolecule #7: Flecainide
| Macromolecule | Name: Flecainide / type: ligand / ID: 7 / Number of copies: 1 / Formula: K4D |
|---|---|
| Molecular weight | Theoretical: 414.343 Da |
| Chemical component information | 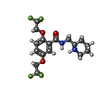 ChemComp-K4D: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.133 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: waiting for 20s, blot for 2.5-3.5s before plunging. | |||||||||
| Details | This sample include channel protein solubilized with GDN detergent. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90.0 K / Max: 103.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Details | Squares on grids were manually screened before data collection |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 5000 / Average exposure time: 8.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.8 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)