+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6uz0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cardiac sodium channel with flecainide | |||||||||
 Components Components | Sodium channel protein type 5 subunit alpha,Green fluorescent protein | |||||||||
 Keywords Keywords | METAL TRANSPORT / Cardiac sodium channel complex with flecainide / ion channel | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated sodium channel activity involved in AV node cell action potential / voltage-gated sodium channel activity involved in bundle of His cell action potential / voltage-gated sodium channel activity involved in Purkinje myocyte action potential / voltage-gated sodium channel activity involved in SA node cell action potential / bundle of His cell action potential / regulation of ventricular cardiac muscle cell membrane depolarization / AV node cell action potential / SA node cell action potential / AV node cell to bundle of His cell communication / membrane depolarization during SA node cell action potential ...voltage-gated sodium channel activity involved in AV node cell action potential / voltage-gated sodium channel activity involved in bundle of His cell action potential / voltage-gated sodium channel activity involved in Purkinje myocyte action potential / voltage-gated sodium channel activity involved in SA node cell action potential / bundle of His cell action potential / regulation of ventricular cardiac muscle cell membrane depolarization / AV node cell action potential / SA node cell action potential / AV node cell to bundle of His cell communication / membrane depolarization during SA node cell action potential / response to denervation involved in regulation of muscle adaptation / sodium channel complex / membrane depolarization during atrial cardiac muscle cell action potential / cardiac ventricle development / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane repolarization / brainstem development / membrane depolarization during AV node cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during bundle of His cell action potential / positive regulation of action potential / membrane depolarization during Purkinje myocyte cell action potential / atrial cardiac muscle cell action potential / telencephalon development / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / ventricular cardiac muscle cell action potential / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / voltage-gated sodium channel complex / sodium ion import across plasma membrane / regulation of cardiac muscle cell contraction / ankyrin binding / voltage-gated sodium channel activity / sodium ion transport / nitric-oxide synthase binding / odontogenesis of dentin-containing tooth / regulation of heart rate by cardiac conduction / fibroblast growth factor binding / intercalated disc / lateral plasma membrane / membrane depolarization / positive regulation of heart rate / neuronal action potential / cardiac muscle contraction / T-tubule / regulation of heart rate / cellular response to calcium ion / sodium ion transmembrane transport / cerebellum development / bioluminescence / positive regulation of epithelial cell proliferation / generation of precursor metabolites and energy / sarcolemma / caveola / Z disc / scaffold protein binding / transmembrane transporter binding / calmodulin binding / protein domain specific binding / axon / ubiquitin protein ligase binding / protein kinase binding / perinuclear region of cytoplasm / enzyme binding / cell surface / endoplasmic reticulum / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Jiang, D. / Shi, H. / Tonggu, L. / Lenaeus, M.J. / Zheng, N. / Catterall, W.A. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Structure of the Cardiac Sodium Channel. Authors: Daohua Jiang / Hui Shi / Lige Tonggu / Tamer M Gamal El-Din / Michael J Lenaeus / Yan Zhao / Craig Yoshioka / Ning Zheng / William A Catterall /  Abstract: Voltage-gated sodium channel Na1.5 generates cardiac action potentials and initiates the heartbeat. Here, we report structures of Na1.5 at 3.2-3.5 Å resolution. Na1.5 is distinguished from other ...Voltage-gated sodium channel Na1.5 generates cardiac action potentials and initiates the heartbeat. Here, we report structures of Na1.5 at 3.2-3.5 Å resolution. Na1.5 is distinguished from other sodium channels by a unique glycosyl moiety and loss of disulfide-bonding capability at the Naβ subunit-interaction sites. The antiarrhythmic drug flecainide specifically targets the central cavity of the pore. The voltage sensors are partially activated, and the fast-inactivation gate is partially closed. Activation of the voltage sensor of Domain III allows binding of the isoleucine-phenylalanine-methionine (IFM) motif to the inactivation-gate receptor. Asp and Ala, in the selectivity motif DEKA, line the walls of the ion-selectivity filter, whereas Glu and Lys are in positions to accept and release Na ions via a charge-delocalization network. Arrhythmia mutation sites undergo large translocations during gating, providing a potential mechanism for pathogenic effects. Our results provide detailed insights into Na1.5 structure, pharmacology, activation, inactivation, ion selectivity, and arrhythmias. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6uz0.cif.gz 6uz0.cif.gz | 253.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6uz0.ent.gz pdb6uz0.ent.gz | 183.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6uz0.json.gz 6uz0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uz/6uz0 https://data.pdbj.org/pub/pdb/validation_reports/uz/6uz0 ftp://data.pdbj.org/pub/pdb/validation_reports/uz/6uz0 ftp://data.pdbj.org/pub/pdb/validation_reports/uz/6uz0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  20949MC  6uz3C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 209037.781 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Gene: Scn5a, GFP / Organ: HEART / Plasmid: pEG_BacMam / Cell line (production host): HEK293S / Production host:  Mammalian 1 orthobornavirus / References: UniProt: P15389, UniProt: P42212 Mammalian 1 orthobornavirus / References: UniProt: P15389, UniProt: P42212 |
|---|
-Sugars , 3 types, 5 molecules 
| #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Source method: isolated from a genetically manipulated source #4: Sugar | |
|---|
-Non-polymers , 3 types, 17 molecules 

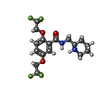


| #5: Chemical | ChemComp-9Z9 / ( #6: Chemical | ChemComp-6OU / [( #7: Chemical | ChemComp-K4D / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: cardiac sodium channel complex with flecainide / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.25 MDa / Experimental value: NO | |||||||||||||||
| Source (natural) | Organism:  | |||||||||||||||
| Source (recombinant) | Organism:  Mammalian 1 orthobornavirus / Strain: HEK293 / Plasmid: pEG_BacMam Mammalian 1 orthobornavirus / Strain: HEK293 / Plasmid: pEG_BacMam | |||||||||||||||
| Buffer solution | pH: 6 | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: This sample include channel protein solubilized with GDN detergent. | |||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil | |||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K / Details: waiting for 20s, blot for 2.5-3.5s before plunging |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS Details: Squares on grids were manually screened before data collection |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Nominal defocus max: 2800 nm / Nominal defocus min: 1000 nm / Calibrated defocus min: 800 nm / Calibrated defocus max: 3000 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Temperature (max): 103 K / Temperature (min): 90 K |
| Image recording | Average exposure time: 8 sec. / Electron dose: 60 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 2 / Num. of real images: 5000 |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
| Image scans | Width: 3838 / Height: 3710 / Movie frames/image: 40 / Used frames/image: 1-40 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.15.2_3472: / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
| Image processing | Details: The images were manually inspected. | ||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2369854 / Details: particles were autopicked in relion3 | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.24 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 513604 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 60 / Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6AGF Pdb chain-ID: A / Accession code: 6AGF / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







