[English] 日本語
 Yorodumi
Yorodumi- EMDB-20872: C3 symmetrical reconstruction of the Staphylococcus aureus phage ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20872 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C3 symmetrical reconstruction of the Staphylococcus aureus phage 80alpha baseplate | |||||||||
 Map data Map data | C3 Tal-focused map used for final model refinement of Tal, Dit, and TMP | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Staphylococcus virus 80alpha Staphylococcus virus 80alpha | |||||||||
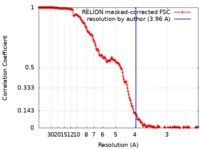
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Kizziah JL / Dokland T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2020 Journal: PLoS Pathog / Year: 2020Title: Structure of the host cell recognition and penetration machinery of a Staphylococcus aureus bacteriophage. Authors: James L Kizziah / Keith A Manning / Altaira D Dearborn / Terje Dokland /  Abstract: Staphylococcus aureus is a common cause of infections in humans. The emergence of virulent, antibiotic-resistant strains of S. aureus is a significant public health concern. Most virulence and ...Staphylococcus aureus is a common cause of infections in humans. The emergence of virulent, antibiotic-resistant strains of S. aureus is a significant public health concern. Most virulence and resistance factors in S. aureus are encoded by mobile genetic elements, and transduction by bacteriophages represents the main mechanism for horizontal gene transfer. The baseplate is a specialized structure at the tip of bacteriophage tails that plays key roles in host recognition, cell wall penetration, and DNA ejection. We have used high-resolution cryo-electron microscopy to determine the structure of the S. aureus bacteriophage 80α baseplate at 3.75 Å resolution, allowing atomic models to be built for most of the major tail and baseplate proteins, including two tail fibers, the receptor binding protein, and part of the tape measure protein. Our structure provides a structural basis for understanding host recognition, cell wall penetration and DNA ejection in viruses infecting Gram-positive bacteria. Comparison to other phages demonstrates the modular design of baseplate proteins, and the adaptations to the host that take place during the evolution of staphylococci and other pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20872.map.gz emd_20872.map.gz | 469.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20872-v30.xml emd-20872-v30.xml emd-20872.xml emd-20872.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20872_fsc.xml emd_20872_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20872.png emd_20872.png | 53.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20872 http://ftp.pdbj.org/pub/emdb/structures/EMD-20872 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20872 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20872 | HTTPS FTP |
-Related structure data
| Related structure data |  6v8iMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20872.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20872.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C3 Tal-focused map used for final model refinement of Tal, Dit, and TMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Staphylococcus phage 80alpha baseplate
| Entire | Name: Staphylococcus phage 80alpha baseplate |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus phage 80alpha baseplate
| Supramolecule | Name: Staphylococcus phage 80alpha baseplate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: 80alpha baseplate attached to fully formed tails with no capsids |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha / Strain: ST247 Staphylococcus virus 80alpha / Strain: ST247 |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 3.7 MDa |
-Macromolecule #1: Major Tail Protein
| Macromolecule | Name: Major Tail Protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MANMKNSNDR IILFRKAGEK VDATKMLFLT EYGLSHEADT DTEDTMDGSY NTGGSVESTM SGTAKMFYGD DFADEIEDAV VDRVLYEAWE VESRIPGKNG DSAKFKAKYF QGFHNKFELK AEANGIDEYE YEYGVNGRFQ RGFATLPEAV TKKLKATGYR FHDTTKEDAL TSEDLTAIPQ PKVDSSTVTP GEV |
-Macromolecule #2: Distal Tail Protein
| Macromolecule | Name: Distal Tail Protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MDIELTKKDG TVIKLSEYGF IVNDIVIDSM QINTKYQDKE NMNGRILMGS NYISRDIVVP CFCKVKNRSD IAYMRDMLYS LTTDIEPMYL REIRRKEELN YRFTQPTSDD YVKLDKNNFP DYEYSRHDQQ IYVNGKQYKV IFNGVINPKQ KGNKVSFELK FETTELPYGE ...String: MDIELTKKDG TVIKLSEYGF IVNDIVIDSM QINTKYQDKE NMNGRILMGS NYISRDIVVP CFCKVKNRSD IAYMRDMLYS LTTDIEPMYL REIRRKEELN YRFTQPTSDD YVKLDKNNFP DYEYSRHDQQ IYVNGKQYKV IFNGVINPKQ KGNKVSFELK FETTELPYGE SIGTSLELEE NKKVGLWSFD FNIDWHAGGD KRKYTFENLS KGTVYYHGSA PNDQFNMYKK ITIILGEDTE SFVWNLTHAE IMKIEGIKLK AGDRIVYDSF RVYKNGVEIS TETNISQPKF KYGANKFEFN QTVQKVQFDL KFYYK |
-Macromolecule #3: Tail-Associated Lysin
| Macromolecule | Name: Tail-Associated Lysin / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MTITIKPPKG NGAPVPVETT LVKKVNADGV LTFDILENKY TYEVINAIGK RWIVSHVEGE NDKKEYVITV IDRKSEGDRQ LVECTAREIP IDKLMIDRIY VNVTGSFTVE RYFNIVFQGT GMLFEVEGKV KSSKFENGGE GDTRLEMFKK GLEHFGLEYK ITYDKKKDRY ...String: MTITIKPPKG NGAPVPVETT LVKKVNADGV LTFDILENKY TYEVINAIGK RWIVSHVEGE NDKKEYVITV IDRKSEGDRQ LVECTAREIP IDKLMIDRIY VNVTGSFTVE RYFNIVFQGT GMLFEVEGKV KSSKFENGGE GDTRLEMFKK GLEHFGLEYK ITYDKKKDRY KFVLTPFANQ KASYFISDEV NANAIKLEED ASDFATFIRG YGNYSGEETF EHAGLVMEAR SALAEIYGDI HAEPFKDGKV TDQETMDKEL QSRLKKSLKQ SLSLDFLVLR ESYPEADPQP GDIVQIKSTK LGLNDLVRIV QVKTIRGINN VIVKQDVTLG EFNREQRYMK KVNTAANYVS GLNDVNLSNP SKAAENLKSK VASIAKSTLD LMSRTDLIED KQQKVSSKTV TTSDGTIVHD FIDKSNIKDV KTIGTIGDSV ARGSHAKTNF TEMLGKKLKA KTTNLARGGA TMATVPIGKE AVENSIYRQA EQIRGDLIIL QGTDDDWLHG YWAGVPIGTD KTDTKTFYGA FCSAIEVIRK NNPASKILVM TATRQCPMSG TMIRRKDTDK NKLGLTLEDY VNAQILACSE LDVPVYDAYH TDYFKPYNPA FRKSSMPDGL HPNERGHEVI MYELIKNYYQ FYG |
-Macromolecule #4: Receptor Binding Protein
| Macromolecule | Name: Receptor Binding Protein / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MDNKLITDLS RVFDYRYVDE NEYNFKLISD MLTDFNFSLE YHRNKEVFAH NGEQIKYEHL NVTSSVSDFL TYLNGRFSNM VLGHNGDGIN EVKDARVDNT GYDHKTLQDR LYHDYSTLDA FTKKVEKAVD ENYKEYRATE YRFEPKEQEP EFITDLSPYT NAVMQSFWVD ...String: MDNKLITDLS RVFDYRYVDE NEYNFKLISD MLTDFNFSLE YHRNKEVFAH NGEQIKYEHL NVTSSVSDFL TYLNGRFSNM VLGHNGDGIN EVKDARVDNT GYDHKTLQDR LYHDYSTLDA FTKKVEKAVD ENYKEYRATE YRFEPKEQEP EFITDLSPYT NAVMQSFWVD PRTKIIYMTQ ARPGNHYMLS RLKPNGQFID RLLVKNGGHG THNAYRYIGN ELWIYSAVLD ANENNKFVRF QYRTGEITYG NEMQDVMPNI FNDRYTSAIY NPIENLMIFR REYKASERQL KNSLNFVEVR SADDIDKGID KVLYQMDIPM EYTSDTQPMQ GITYDAGILY WYTGDSKPAN PNYLQGFDIK TKELLFKRRI DIGGVNNNFK GDFQEAEGLD MYYDLETGRK ALLIGVTIGP GNNRHHSIYS IGQRGVNQFL KNIAPQVSMT DSGGRVKPLP IQNPAYLSDI TEVGHYYIYT QDTQNALDFP LPKAFRDAGW FFDVLPGHYN GALRQVLTRN STGRNMLKFE RVIDIFNKKN NGAWNFCPQN AGYWEHIPKS ITKLSDLKIV GLDFYITTEE SNRFTDFPKD FKGIAGWILE VKSNTPGNTT QVLRRNNFPS AHQFLVRNFG TGGVGKWSLF EGKVVE |
-Macromolecule #5: Fiber Lower
| Macromolecule | Name: Fiber Lower / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MVVDNFSKDD NLIELQTTSQ YNPVIDTNIS FYESDRGTGV LNFAVTKNNR PLSISSEHVK TFIVLKTDDY NVDRGAYISD ELTIVDAING RLQYVIPNEF LKHSGKVHAQ AFFTQNGSDN VVVERQFSFN IENDLVSGFD GITKLVYIKS IQDTIEAVGK DFNQLKQNMA ...String: MVVDNFSKDD NLIELQTTSQ YNPVIDTNIS FYESDRGTGV LNFAVTKNNR PLSISSEHVK TFIVLKTDDY NVDRGAYISD ELTIVDAING RLQYVIPNEF LKHSGKVHAQ AFFTQNGSDN VVVERQFSFN IENDLVSGFD GITKLVYIKS IQDTIEAVGK DFNQLKQNMA DTQTLIAKVN DSATKGIQQI EIKQNEAIQA ITATQTSATQ AVTAEFDKIV EKEQAIFERV NEVEQQINGA DLVKGNSTTN WQKSKLTDDY GKAIESYEQS IDSVLSAVNT SRIIHITSAT DAPSFKDIGT VDTPKEDGVD DGSDIPVAPN TLGKSGVLVV YVVDDSTARA TWYPDDSNDE YTKYKISGTW YPFYKKNDGD LTKQFVEETS NNALNQAKQY VDDKFGTTSW QQHKMTEANG QSIQVNLNNA QGDLGYLTAG NYYATRVPDL PGSVESYEGY LSVFVKDDTN KLFNFTPYNS KKIYTRSITN GRLEQQWTVP NEHKSTVLFD GGANGVGTTI NLTEPYTNYS ILLVSGTYPG GVIEGFGLTA LPNAIQLSKA NVVDSDGNGG GIYECLLSKT SSTTLRIDND VYFDLGKTSG SGANANKVTI TKIMGWK |
-Macromolecule #6: Fiber Upper
| Macromolecule | Name: Fiber Upper / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MYKIKDVETR IKNDGVDLGD IGCRFYTEDE NTASIRIGIN DKQGRIDLKA HGLTPRLHLF MEDGSIFKNE PLIIDDVVKG FITYKIPKKV IKHAGYVRCK LFLEKEEEKI HVANFSFNIV DSGIESAVAK EIDVKLVDDA ITRILKDNAT DLLSKDFKEK IDKDVISYIE ...String: MYKIKDVETR IKNDGVDLGD IGCRFYTEDE NTASIRIGIN DKQGRIDLKA HGLTPRLHLF MEDGSIFKNE PLIIDDVVKG FITYKIPKKV IKHAGYVRCK LFLEKEEEKI HVANFSFNIV DSGIESAVAK EIDVKLVDDA ITRILKDNAT DLLSKDFKEK IDKDVISYIE KNESRFKGAK GDKGEPGQPG AKGEAGKKGE QGAPGKNGTV VSINPDTKMW QIDGKDTDIK AEPELLDKIN IANVEGLEDK LQEVEKIKDT TLNDSKTYTD TKIAELVDSA PESMNTLREL AEAIQNNSIS ESVLQQIGSK VSTEDFEEFK QTLNDLYAPK NHNHDERYVL SSQAFTKQQA DSLYQLKSAS QPTVKIWTGT ENEYNYIYQK DPNTLYLIKG |
-Macromolecule #7: Tape Measure Protein
| Macromolecule | Name: Tape Measure Protein / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus virus 80alpha Staphylococcus virus 80alpha |
| Recombinant expression | Organism:  |
| Sequence | String: MTEYKIKATI EASVAKFKRQ IDSAVKSVQR FKRVADQTKD VELNANDKKL QKTIKVAKKS LDAFSNKNVK AKLDASIQDL QQKILESNFE LDKLNSKEAS PEVKLQKQKL TKDIAEAENK LSELEKKRVN IDVNADNSKF NRVLKVSKAS LEALNRSKAK AILDVDNSVA ...String: MTEYKIKATI EASVAKFKRQ IDSAVKSVQR FKRVADQTKD VELNANDKKL QKTIKVAKKS LDAFSNKNVK AKLDASIQDL QQKILESNFE LDKLNSKEAS PEVKLQKQKL TKDIAEAENK LSELEKKRVN IDVNADNSKF NRVLKVSKAS LEALNRSKAK AILDVDNSVA NSKIKRTKEE LKSIPNKTRS RLDVDTRLSI PTIYAFKKSL DALPNKKTTK VDVDTNGLKK VYAYIIKAND NFQRQMGNLA NMFRVFGTVG SNMVGGLLTS SFSILIPVIA SVVPVVFALL NAIKVLTGGV LALGGAVAIA GAGFVAFGAM AISAIKMLND GTLQASSATN EYKKALDGVK SAWTDIIKQN QSAIFTTLAN GLNTVKTAMQ SLQPFFSGIS RGMEEASQSV LKWAENSSVA SRFFNMMNTT GVSVFNKLLS AAGGFGDGLV NVFTQLAPLF QWSADWLDRL GQSFSNWANS AAGENSITRF IEYTKTNLPI IGNIFKNVFA GINNLMNAFS GSSTGIFQSL EQMTAKFREW SEQVGQSQGF KDFVSYIQTN GPLIMQLIGN IARGLVAFAT AMAPIASAVL RVAVAITGWI ANLFEAHPAT AQLVGVIITL VGAFRFLIAP ILAVMDFLGP LAARLVALVT KFGWAKTGTL VLSKAMTSLK GPIKLVTAIF QLLFGKIGLI RNAITGLVTV FGILGGPITI VIGVIAALIA IFVLLWNKNE GFRNFIINAW NAIKTFMVNV WNVLKAVASV VWNAILTAIT TAVSNVYNFI MIVWNQIVAY LQGLWNGIIA IATTVWNLLV TIITTVFTTI MTIVMTIWTA IWTFLSTIWN TIITIATTIW NLLVTVITTV FTTIMTIAMT IWNAIWTFLQ TLWNTIVTVA TKVWNAITTA ISTALQAAWS FISNIWNTIW SFLSGILTTI WNKVVSIFTQ VVSTISDKMS QAWNFIVTKG MQWVSTITST LINFVNRVIQ GFVNVVNKVS QGMTNAVNKI KSFIGDFVSA GADMIRGLIR GIGQMAGQLV DAAKNVAKKA LDAAKSALGI HSPSREFMDV GMYSMLGFVK GIDNHSSKVI RNVSNVADKV VDAFQPTLNA PDISSITGNL SNLGGNINAQ VQHTHSIETS PNMKTVKVEF DVNNDALTSI VNGRNAKRNS EYYL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: NICKEL / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | 80alpha tails (with baseplates) purified by centrifugation, polyethylene glycol precipitation, and CsCl density gradation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Digitization - Frames/image: 1-37 / Number grids imaged: 10 / Number real images: 6483 / Average exposure time: 1.156 sec. / Average electron dose: 104.33 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.7 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)