[English] 日本語
 Yorodumi
Yorodumi- EMDB-20866: Cryo-EM structure of the Thermococcus gammatolerans McrBC complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20866 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Thermococcus gammatolerans McrBC complex | |||||||||
 Map data Map data | FL-TgMcrBC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Endonuclease / AAA protein / GTPase / Methylation-dependent restriction / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationendonuclease activity / ATP hydrolysis activity / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Thermococcus gammatolerans (archaea) Thermococcus gammatolerans (archaea) | |||||||||
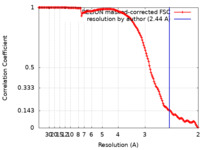
| Method | single particle reconstruction / cryo EM / Resolution: 2.44 Å | |||||||||
 Authors Authors | Niu Y / Suzuki H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes. Authors: Yiming Niu / Hiroshi Suzuki / Christopher J Hosford / Thomas Walz / Joshua S Chappie /   Abstract: McrBC complexes are motor-driven nucleases functioning in bacterial self-defense by cleaving foreign DNA. The GTP-specific AAA + protein McrB powers translocation along DNA and its hydrolysis ...McrBC complexes are motor-driven nucleases functioning in bacterial self-defense by cleaving foreign DNA. The GTP-specific AAA + protein McrB powers translocation along DNA and its hydrolysis activity is stimulated by its partner nuclease McrC. Here, we report cryo-EM structures of Thermococcus gammatolerans McrB and McrBC, and E. coli McrBC. The McrB hexamers, containing the necessary catalytic machinery for basal GTP hydrolysis, are intrinsically asymmetric. This asymmetry directs McrC binding so that it engages a single active site, where it then uses an arginine/lysine-mediated hydrogen-bonding network to reposition the asparagine in the McrB signature motif for optimal catalytic function. While the two McrBC complexes use different DNA-binding domains, these contribute to the same general GTP-recognition mechanism employed by all G proteins. Asymmetry also induces distinct inter-subunit interactions around the ring, suggesting a coordinated and directional GTP-hydrolysis cycle. Our data provide insights into the conserved molecular mechanisms governing McrB family AAA + motors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20866.map.gz emd_20866.map.gz | 228.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20866-v30.xml emd-20866-v30.xml emd-20866.xml emd-20866.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20866_fsc.xml emd_20866_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_20866.png emd_20866.png | 76.8 KB | ||
| Masks |  emd_20866_msk_1.map emd_20866_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20866.cif.gz emd-20866.cif.gz | 6.6 KB | ||
| Others |  emd_20866_half_map_1.map.gz emd_20866_half_map_1.map.gz emd_20866_half_map_2.map.gz emd_20866_half_map_2.map.gz | 195.4 MB 195.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20866 http://ftp.pdbj.org/pub/emdb/structures/EMD-20866 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20866 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20866 | HTTPS FTP |
-Related structure data
| Related structure data |  6ut5MC  6ut3C  6ut4C  6ut6C  6ut7C  6ut8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10583 (Title: Single-particle Cryo-EM of the Thermococcus gammatolerans McrBC complex EMPIAR-10583 (Title: Single-particle Cryo-EM of the Thermococcus gammatolerans McrBC complexData size: 883.3 Data #1: Unaligned multi-frame micrographs of the Thermococcus gammatolerans McrBC complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20866.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20866.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FL-TgMcrBC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20866_msk_1.map emd_20866_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_20866_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_20866_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the full-length Thermococcus gammatolerans McrBC
| Entire | Name: Complex of the full-length Thermococcus gammatolerans McrBC |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the full-length Thermococcus gammatolerans McrBC
| Supramolecule | Name: Complex of the full-length Thermococcus gammatolerans McrBC type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Thermococcus gammatolerans (archaea) Thermococcus gammatolerans (archaea) |
-Macromolecule #1: GTPase subunit of restriction endonuclease
| Macromolecule | Name: GTPase subunit of restriction endonuclease / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus gammatolerans (archaea) Thermococcus gammatolerans (archaea) |
| Molecular weight | Theoretical: 71.668867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MENQLFIIGI GTGTDEYENF EETILKGVKR NELEGQIGPD ILDNCCSDVC YFWGRSKETI YEKKIDKGDM VLFYVGKRIS RNKVDLNQE TAVYLGIICE TVEISENDVS FLNDFWRKGE NFRFLMFFKK KPEKLHHSIN EINSKLGYNP DYFPIAGYVK P ERMSGVYD ...String: MENQLFIIGI GTGTDEYENF EETILKGVKR NELEGQIGPD ILDNCCSDVC YFWGRSKETI YEKKIDKGDM VLFYVGKRIS RNKVDLNQE TAVYLGIICE TVEISENDVS FLNDFWRKGE NFRFLMFFKK KPEKLHHSIN EINSKLGYNP DYFPIAGYVK P ERMSGVYD ILKNILKKRG ILKESDSMNE SAGHNIKEDY FRVDMLLNKK GQVILYGPPG TGKTWIARKY VVEETNEKTP GN KWEFITF HQSYSYEEFI EGFRPRTDNE EKIRYVVEDG IFKKIALRAL VKGLFELEDA TIGKDKIHRL YILLTKKEPL SPT EYEEYL RLKRYLWELV GGLPKDKLKN LTPKFYLIID EINRGNISKI FGELITLLEK DKRLGGENQL IVRLPYSGEP FAVP PNLYI IGTMNTADRS IALLDVALRR RFAFIEVEPR PEFLEKENLK KIREKKLKTE DRKRLNEKLN ELFSKLGNDN YFLKT LLEK INVRITVVKD RDHRIGHSYF LNVETVEDLH HVWYYEVLPL LMEYFYNDWE TIKWVLNEKG KEHGNVFFEK LRLTGP NGE EAYQLKVLEG DAFIGALKRI ISKNTPSQEG GATTNEENSP ENTQSQTEGD UniProtKB: GTPase subunit of restriction endonuclease |
-Macromolecule #2: McrBC 5-methylcytosine restriction system component
| Macromolecule | Name: McrBC 5-methylcytosine restriction system component / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus gammatolerans (archaea) Thermococcus gammatolerans (archaea) |
| Molecular weight | Theoretical: 54.339406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPRLTTITLY EHDEKRYRDI AGDKKAIQDA LIKLNKQFKK DFKKLDRSED NSDTEDTIDE SKGVVEVYAN KIKARHYVGF AAVDNVFLQ ILPKVFKPKK EQTQETQEDT WEPILAFIRM LDMAYGLKIK DHDLAYLQGR NLRPNLYEVF IYLFAKSLWS E VQRGYHRE ...String: MPRLTTITLY EHDEKRYRDI AGDKKAIQDA LIKLNKQFKK DFKKLDRSED NSDTEDTIDE SKGVVEVYAN KIKARHYVGF AAVDNVFLQ ILPKVFKPKK EQTQETQEDT WEPILAFIRM LDMAYGLKIK DHDLAYLQGR NLRPNLYEVF IYLFAKSLWS E VQRGYHRE YVEVHREEKF LRGKLLMSRQ IRKLPHQLNT FSVEVHELIE DNLLNRIFYA SVREALRRTT WGLNRKLLGE LM LAFDGIT PIHLRTEHFE RVHFTRLNER FRRPFELAKL LFMPASGKGR SREVSGFFVD MNKLFERFIE RVLVRNLPPE YKL FYQESY PFLKNQNGSS QKPDYVVRKG NTPVVVLDAK YRELKERIPS SDMLRQLYVY SRIWGYKTSH ENDSKPPAVI VIPS SSTYN QGLPDKPLEF EFFDERKLFI VAYNMDYVKT GAIFKADKNF RRSLNNIIGK LNT UniProtKB: McrBC 5-methylcytosine restriction system component |
-Macromolecule #3: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE
| Macromolecule | Name: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: GSP |
|---|---|
| Molecular weight | Theoretical: 539.246 Da |
| Chemical component information |  ChemComp-GSP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 28 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 16.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2078 / Average exposure time: 10.0 sec. / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)