[English] 日本語
 Yorodumi
Yorodumi- EMDB-20826: In situ structure of Parkinson's disease-linked Microtubule-Bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20826 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
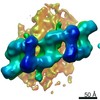
| Title | In situ structure of Parkinson's disease-linked Microtubule-Bound human LRRK2(I2020T) MASK B | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Boehning J / Buschauer R / Watanabe R / Villa E | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: The In Situ Structure of Parkinson's Disease-Linked LRRK2. Authors: Reika Watanabe / Robert Buschauer / Jan Böhning / Martina Audagnotto / Keren Lasker / Tsan-Wen Lu / Daniela Boassa / Susan Taylor / Elizabeth Villa /  Abstract: Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most frequent cause of familial Parkinson's disease. LRRK2 is a multi-domain protein containing a kinase and GTPase. Using correlative light ...Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most frequent cause of familial Parkinson's disease. LRRK2 is a multi-domain protein containing a kinase and GTPase. Using correlative light and electron microscopy, in situ cryo-electron tomography, and subtomogram analysis, we reveal a 14-Å structure of LRRK2 bearing a pathogenic mutation that oligomerizes as a right-handed double helix around microtubules, which are left-handed. Using integrative modeling, we determine the architecture of LRRK2, showing that the GTPase and kinase are in close proximity, with the GTPase closer to the microtubule surface, whereas the kinase is exposed to the cytoplasm. We identify two oligomerization interfaces mediated by non-catalytic domains. Mutation of one of these abolishes LRRK2 microtubule-association. Our work demonstrates the power of cryo-electron tomography to generate models of previously unsolved structures in their cellular environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20826.map.gz emd_20826.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20826-v30.xml emd-20826-v30.xml emd-20826.xml emd-20826.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20826.png emd_20826.png | 100.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20826 http://ftp.pdbj.org/pub/emdb/structures/EMD-20826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20826 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20826.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20826.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : FIB-milled cellular samples expressing Parkinson's disease mutant...
| Entire | Name: FIB-milled cellular samples expressing Parkinson's disease mutant LRRK2 (I2020T) in HEK293 cells |
|---|---|
| Components |
|
-Supramolecule #1: FIB-milled cellular samples expressing Parkinson's disease mutant...
| Supramolecule | Name: FIB-milled cellular samples expressing Parkinson's disease mutant LRRK2 (I2020T) in HEK293 cells type: cell / ID: 1 / Parent: 0 / Macromolecule list: all Details: FIB-milled cellular samples expressing Parkinson's disease-related mutant LRRK2 (I2020T) proteins in human embryonic kidney cells (HEK293 cells) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Cell: HEK293 Homo sapiens (human) / Cell: HEK293 |
-Macromolecule #1: Leucine Rich Repeat Kinase 2
| Macromolecule | Name: Leucine Rich Repeat Kinase 2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASGSCQGCE EDEETLKKLI VRLNNVQEGK QIETLVQILE DLLVFTYSER ASKLFQGKNI HVPLLIVLD SYMRVASVQQ VGWSLLCKLI EVCPGTMQSL MGPQDVGNDW EVLGVHQLIL K MLTVHNAS VNLSVIGLKT LDLLLTSGKI TLLILDEESD IFMLIFDAMH ...String: MASGSCQGCE EDEETLKKLI VRLNNVQEGK QIETLVQILE DLLVFTYSER ASKLFQGKNI HVPLLIVLD SYMRVASVQQ VGWSLLCKLI EVCPGTMQSL MGPQDVGNDW EVLGVHQLIL K MLTVHNAS VNLSVIGLKT LDLLLTSGKI TLLILDEESD IFMLIFDAMH SFPANDEVQK LG CKALHVL FERVSEEQLT EFVENKDYMI LLSALTNFKD EEEIVLHVLH CLHSLAIPCN NVE VLMSGN VRCYNIVVEA MKAFPMSERI QEVSCCLLHR LTLGNFFNIL VLNEVHEFVV KAVQ QYPEN AALQISALSC LALLTETIFL NQDLEEKNEN QENDDEGEED KLFWLEACYK ALTWH RKNK HVQEAACWAL NNLLMYQNSL HEKIGDEDGH FPAHREVMLS MLMHSSSKEV FQASAN ALS TLLEQNVNFR KILLSKGIHL NVLELMQKHI HSPEVAESGC KMLNHLFEGS NTSLDIM AA VVPKILTVMK RHETSLPVQL EALRAILHFI VPGMPEESRE DTEFHHKLNM VKKQCFKN D IHKLVLAALN RFIGNPGIQK CGLKVISSIV HFPDALEMLS LEGAMDSVLH TLQMYPDDQ EIQCLGLSLI GYLITKKNVF IGTGHLLAKI LVSSLYRFKD VAEIQTKGFQ TILAILKLSA SFSKLLVHH SFDLVIFHQM SSNIMEQKDQ QFLNLCCKCF AKVAMDDYLK NVMLERACDQ N NSIMVECL LLLGADANQA KEGSSLICQV CEKESSPKLV ELLLNSGSRE QDVRKALTIS IG KGDSQII SLLLRRLALD VANNSICLGG FCIGKVEPSW LGPLFPDKTS NLRKQTNIAS TLA RMVIRY QMKSAVEEGT ASGSDGNFSE DVLSKFDEWT FIPDSSMDSV FAQSDDLDSE GSEG SFLVK KKSNSISVGE FYRDAVLQRC SPNLQRHSNS LGPIFDHEDL LKRKRKILSS DDSLR SSKL QSHMRHSDSI SSLASEREYI TSLDLSANEL RDIDALSQKC CISVHLEHLE KLELHQ NAL TSFPQQLCET LKSLTHLDLH SNKFTSFPSY LLKMSCIANL DVSRNDIGPS VVLDPTV KC PTLKQFNLSY NQLSFVPENL TDVVEKLEQL ILEGNKISGI CSPLRLKELK ILNLSKNH I SSLSENFLEA CPKVESFSAR MNFLAAMPFL PPSMTILKLS QNKFSCIPEA ILNLPHLRS LDMSSNDIQY LPGPAHWKSL NLRELLFSHN QISILDLSEK AYLWSRVEKL HLSHNKLKEI PPEIGCLEN LTSLDVSYNL ELRSFPNEMG KLSKIWDLPL DELHLNFDFK HIGCKAKDII R FLQQRLKK AVPYNRMKLM IVGNTGSGKT TLLQQLMKTK KSDLGMQSAT VGIDVKDWPI QI RDKRKRD LVLNVWDFAG REEFYSTHPH FMTQRALYLA VYDLSKGQAE VDAMKPWLFN IKA RASSSP VILVGTHLDV SDEKQRKACM SKITKELLNK RGFPAIRDYH FVNATEESDA LAKL RKTII NESLNFKIRD QLVVGQLIPD CYVELEKIIL SERKNVPIEF PVIDRKRLLQ LVREN QLQL DENELPHAVH FLNESGVLLH FQDPALQLSD LYFVEPKWLC KIMAQILTVK VEGCPK HPK GIISRRDVEK FLSKKRKFPK NYMSQYFKLL EKFQIALPIG EEYLLVPSSL SDHRPVI EL PHCENSEIII RLYEMPYFPM GFWSRLINRL LEISPYMLSG RERALRPNRM YWRQGIYL N WSPEAYCLVG SEVLDNHPES FLKITVPSCR KGCILLGQVV DHIDSLMEEW FPGLLEIDI CGEGETLLKK WALYSFNDGE EHQKILLDDL MKKAEEGDLL VNPDQPRLTI PISQIAPDLI LADLPRNIM LNNDELEFEQ APEFLLGDGS FGSVYRAAYE GEEVAVKIFN KHTSLRLLRQ E LVVLCHLH HPSLISLLAA GIRPRMLVME LASKGSLDRL LQQDKASLTR TLQHRIALHV AD GLRYLHS AMIIYRDLKP HNVLLFTLYP NAAIIAKIAD YGTAQYCCRM GIKTSEGTPG FRA PEVARG NVIYNQQADV YSFGLLLYDI LTTGGRIVEG LKFPNEFDEL EIQGKLPDPV KEYG CAPWP MVEKLIKQCL KENPQERPTS AQVFDILNSA ELVCLTRRIL LPKNVIVECM VATHH NSRN ASIWLGCGHT DRGQLSFLDL NTEGYTSEEV ADSRILCLAL VHLPVEKESW IVSGTQ SGT LLVINTEDGK KRHTLEKMTD SVTCLYCNSF SKQSKQKNFL LVGTADGKLA IFEDKTV KL KGAAPLKILN IGNVSTPLMC LSESTNSTER NVMWGGCGTK IFSFSNDFTI QKLIETRT S QLFSYAAFSD SNIITVVVDT ALYIAKQNSP VVEVWDKKTE KLCGLIDCVH FLREVMVKE NKESKHKMSY SGRVKTLCLQ KNTALWIGTG GGHILLLDLS TRRLIRVIYN FCNSVRVMMT AQLGSLKNV MLVLGYNRKN TEGTQKQKEI QSCLTVWDIN LPHEVQNLEK HIEVRKELAE K MRRTSVE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Details: unspecified |
| Vitrification | Cryogen name: ETHANE-PROPANE |
| Details | FIB-milled cellular samples expressing Parkinson's disease-related mutant LRRK2 (I2020T) proteins in human embryonic kidney cells (HEK293 cells) |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number subtomograms used: 4307 |
|---|---|
| Extraction | Number tomograms: 12 / Number images used: 11508 |
| CTF correction | Software - Name: CTFFIND |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















