[English] 日本語
 Yorodumi
Yorodumi- EMDB-20557: Atomic structure of the Human Herpesvirus 6B Capsid and Capsid-As... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20557 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic structure of the Human Herpesvirus 6B Capsid and Capsid-Associated Tegument Complexes | |||||||||||||||
 Map data Map data | HHV-6B capsid associated with pU11 tegument proteins | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | beta-herpesvirus / HHV-6B / murine cytomegalovirus / human cytomegalovirus / pp150 / pU11 / pUL32 / pM32 / pU14 / VIRUS | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=16 icosahedral viral capsid / viral tegument / viral capsid assembly / viral process / viral capsid / host cell nucleus / structural molecule activity / DNA binding Similarity search - Function | |||||||||||||||
| Biological species |  Human herpesvirus 6B (strain Z29) / Human herpesvirus 6B (strain Z29) /  Human herpesvirus 6 strain Z29 Human herpesvirus 6 strain Z29 | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||||||||
 Authors Authors | Zhang YB / Liu W | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes. Authors: Yibo Zhang / Wei Liu / Zihang Li / Vinay Kumar / Ana L Alvarez-Cabrera / Emily C Leibovitch / Yanxiang Cui / Ye Mei / Guo-Qiang Bi / Steve Jacobson / Z Hong Zhou /   Abstract: Human herpesvirus 6B (HHV-6B) belongs to the β-herpesvirus subfamily of the Herpesviridae. To understand capsid assembly and capsid-tegument interactions, here we report atomic structures of HHV-6B ...Human herpesvirus 6B (HHV-6B) belongs to the β-herpesvirus subfamily of the Herpesviridae. To understand capsid assembly and capsid-tegument interactions, here we report atomic structures of HHV-6B capsid and capsid-associated tegument complex (CATC) obtained by cryoEM and sub-particle reconstruction. Compared to other β-herpesviruses, HHV-6B exhibits high similarity in capsid structure but organizational differences in its CATC (pU11 tetramer). 180 "VΛ"-shaped CATCs are observed in HHV-6B, distinguishing from the 255 "Λ"-shaped dimeric CATCs observed in murine cytomegalovirus and the 310 "Δ"-shaped CATCs in human cytomegalovirus. This trend in CATC quantity correlates with the increasing genomes sizes of these β-herpesviruses. Incompatible distances revealed by the atomic structures rationalize the lack of CATC's binding to triplexes Ta, Tc, and Tf in HHV-6B. Our results offer insights into HHV-6B capsid assembly and the roles of its tegument proteins, including not only the β-herpesvirus-specific pU11 and pU14, but also those conserved across all subfamilies of Herpesviridae. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20557.map.gz emd_20557.map.gz | 911.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20557-v30.xml emd-20557-v30.xml emd-20557.xml emd-20557.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20557.png emd_20557.png | 188.5 KB | ||
| Filedesc metadata |  emd-20557.cif.gz emd-20557.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20557 http://ftp.pdbj.org/pub/emdb/structures/EMD-20557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20557 | HTTPS FTP |
-Related structure data
| Related structure data |  6q1fMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20557.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20557.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HHV-6B capsid associated with pU11 tegument proteins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.17 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
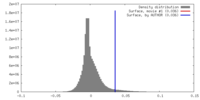
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human herpesvirus 6 strain Z29
| Entire | Name:  Human herpesvirus 6 strain Z29 Human herpesvirus 6 strain Z29 |
|---|---|
| Components |
|
-Supramolecule #1: Human herpesvirus 6 strain Z29
| Supramolecule | Name: Human herpesvirus 6 strain Z29 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 36351 / Sci species name: Human herpesvirus 6 strain Z29 / Sci species strain: Z29 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 6B (strain Z29) / Strain: Z29 Human herpesvirus 6B (strain Z29) / Strain: Z29 |
| Molecular weight | Theoretical: 152.260047 KDa |
| Sequence | String: MENWQATEIL PKIEAPLNIF NDIKTYTAEQ LFDNLRIYFG DDPSRYNISF EALLGIYCNK IEWINFFTTP IAVAANVIRF NDVSRMTLG KVLFFIQLPR VATGNDVTAP KETTIMVAKH SEKHPINISF DLSAACLEHL ENTFKNTVID QILNINALHT V LRSLKNSA ...String: MENWQATEIL PKIEAPLNIF NDIKTYTAEQ LFDNLRIYFG DDPSRYNISF EALLGIYCNK IEWINFFTTP IAVAANVIRF NDVSRMTLG KVLFFIQLPR VATGNDVTAP KETTIMVAKH SEKHPINISF DLSAACLEHL ENTFKNTVID QILNINALHT V LRSLKNSA DSLERGLIHA FMQTLLRKSP PQFIVLTMNE NKVHNKQALS RVQRSNMFQS LKNRLLTSLF FLNRNNNSSY IY RILNDMM ESVTESILND TNNYTSKENI PLDGVLLGPI GSIQKLTNIL SQYISTQVVS APISYGHFIM GKENAVTAIA YRA IMADFT QFTVNAGTEQ QDTNNKSEIF DKSRAYADLK LNTLKLGDKL VAFDHLHKVY KNTDVNDPLE QSLQLTFFFP LGIY IPTET GFSTMETRVK LNDTMENNLP TSVFFHNKDQ VVQRIDFADI LPSVCHPIVH DSTIVERLMK NEPLPTGHRF SQLCQ LKIT RENPTRILQT LYNLYESRQE VPKNTNVLKN ELNVEDFYKP DNPTLPTERH PFFDLTYIQK NRATEVLCTP RIMIGN MPL PLAPISFHEA RTNQMLEHAK TNSHNYDFTL KIVTESLTSG SYPELAYVIE ILVHGNKHAF MILKQVISQC ISYWFNM KH ILLFCNSFEM IMLISNHMGD ELIPGAAFAH YRNLVSLIRL VKRTISISNI NEQLCGEPLV NFANALFDGR LFCPFVHT M PRNDTNAKIT ADDTPLTQNT VRVRNYEISD VQRMNLIDSS VVFTDNDRPS NENTILSKIF YFCVLPALSN NKACGAGVN VKELVLDLFY TEPFICPDDC FQENPISSDV LMSLIREAMG PGYTVANTSS IAKQLFKSLI YINENTKILE VEVSLDPAQR HGNSVHFQS LQHILYNGLC LISPITTLRR YYQPIPFHRF FSDPGICGTM NADIQVFLNT FPHYQRNDGG FPLPPPLALE F YNWQRTPF SVYSAFCPNS LLSIMTLAAM HSKLSPVAIA IQSKSKIHPG FAATLVRTDN FDVECLLYSS RAATSIILDD PT VTAEAKD IVTTYNFTQH LSFVDMGLGF SSTTATANLK RIKSDMGSKI QNLFSAFPIH AFTNTDINTW IRHHVGIEKP NPS EGEALN IITFGGINKN PPSILLHGQQ AICEVILTPV TTNINFFKLP HNPRGRESCM MGTDPHNEEA ARKALYDHTQ TDSD TFAAT TNPWASLPGS LGDILYNTAH REQLCYNPKT YSPNAQFFTE SDILKTNKMM YKVINEYCMK SNSCLNSDSE IQYSC SEGT DSFVSRPCQF LQNALPLHCS SNQALLESRS KTGNTQISET HYCNYAIGET IPLQLIIESS I UniProtKB: Major capsid protein |
-Macromolecule #2: Large structural phosphoprotein
| Macromolecule | Name: Large structural phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 6B (strain Z29) / Strain: Z29 Human herpesvirus 6B (strain Z29) / Strain: Z29 |
| Molecular weight | Theoretical: 95.738125 KDa |
| Sequence | String: MDLKAQSIPF AWLDRDKVQR LTNFLSNLEN LENVDLREHP YVTNSCVVRE GEDVDELKTL YNTFILWLMY HYVLSKRKPD YNAIWQDIT KLQNVVNEYL KSKGLNKGNF ENMFTNKEKF ESQFSDIHRA LLRLGNSIRW GSNVPIDTPY VNLTAEDSSE I ENNLQDAE ...String: MDLKAQSIPF AWLDRDKVQR LTNFLSNLEN LENVDLREHP YVTNSCVVRE GEDVDELKTL YNTFILWLMY HYVLSKRKPD YNAIWQDIT KLQNVVNEYL KSKGLNKGNF ENMFTNKEKF ESQFSDIHRA LLRLGNSIRW GSNVPIDTPY VNLTAEDSSE I ENNLQDAE KNMLWYTVYN INDPWDENGY LVTSINKLVY LGKLFVTLNQ SWSKLEKVAM SQIVTTQNHL SGHLRKNENF NA VYSQRVL QTPLTGQRVE SFLKIITSDY EIIKSSLESY SASKAFSVPE NGPHSLMDFA SLDGRMPSDL SLPSISIDTK RPS ADLARL KISQPKSLDA PLKTQRRHKF PESDSVDNAG GKILIKKETL GGRDVRATTP VSSVSLMSGV EPLSSLTSTN LDLR DKSHG NYRIGPSGIL DFGVKLPAEA QSNTGDVDLL QDKTSIRSPS SGITDVVNGL ANLNLRQNKS DVSRPWSKNT AANAD VFDP VHRLVSEQTG TPFVLNNSDV AGSEAKLTTH STETGVSPHN VSLIKDLRDK DGFRKQKKLD LLGSWTKEKN DKAIVH SRE VTGDSGDATE TVTARDSPVL RKTKHANDIF AGLNKKYARD VSRGGKGNSR DLYSGGNAEK KETSGKFNVD KEMTQNE QE PLPNLMEAAR NAGEEQYVQA GLGQRVNKIL AEFTNLISLG EKGIQDILHN QSGTELKLPT ENKLGRESEE ANVERILE V SDPQNLFKNF KLQNDLDSVQ SPFRLPNADL SRDLDSVSFK DALDVKLPGN GEREIDLALQ KVKAGERETS DFKVGQDET LIPTQLMKVE TPEEKDDVIE KMVLRIRQDG ETDEETVPGP GVAESLGIAA KDKSVIAS UniProtKB: Large structural phosphoprotein |
-Macromolecule #3: Small capsomere-interacting protein
| Macromolecule | Name: Small capsomere-interacting protein / type: protein_or_peptide / ID: 3 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 6B (strain Z29) / Strain: Z29 Human herpesvirus 6B (strain Z29) / Strain: Z29 |
| Molecular weight | Theoretical: 9.827329 KDa |
| Sequence | String: MTTIRGDDLS NQITQISGSS SKKEEEKKKQ QMLTGVLGLQ PTMANHPVLG VFLPKYAKQN GGNVDKTAFR LDLIRMLALH RLNTKTGSD UniProtKB: Small capsomere-interacting protein |
-Macromolecule #4: Triplex capsid protein 1
| Macromolecule | Name: Triplex capsid protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 6B (strain Z29) / Strain: Z29 Human herpesvirus 6B (strain Z29) / Strain: Z29 |
| Molecular weight | Theoretical: 34.162508 KDa |
| Sequence | String: MNSKSSARAA IVDTVEAVKK RKYISIEAGT LNNVVEKERK FLKQFLSGRE NLRIAARVFT PCELLAPELE NLGMLMYRFE TDVDNPKIL FVGLFFLCSN AFNVSACVRT ALTTMYTNSM VDNVLSMINT CKYLEDKVSL FGVTSLVSCG SSCLLSCVMQ G NVYDANKE ...String: MNSKSSARAA IVDTVEAVKK RKYISIEAGT LNNVVEKERK FLKQFLSGRE NLRIAARVFT PCELLAPELE NLGMLMYRFE TDVDNPKIL FVGLFFLCSN AFNVSACVRT ALTTMYTNSM VDNVLSMINT CKYLEDKVSL FGVTSLVSCG SSCLLSCVMQ G NVYDANKE NIHGLTVLKE IFLEPDWEPR QHSTQYVYVV HVYKEVLSKL QYGIYVVLTS FQNEDLVVDI LRQYFEKERF LF LNYLINS NTTLSYFGSV QRIGRCATED IKSGFLQYRG ITLPVIKLEN IFVDLSEKKV FV UniProtKB: Triplex capsid protein 1 |
-Macromolecule #5: Triplex capsid protein 2
| Macromolecule | Name: Triplex capsid protein 2 / type: protein_or_peptide / ID: 5 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 6B (strain Z29) / Strain: Z29 Human herpesvirus 6B (strain Z29) / Strain: Z29 |
| Molecular weight | Theoretical: 33.514332 KDa |
| Sequence | String: METVYCTFDH KLSLSDISTL CKLMNIVIPI PAHHHLIGSG NLGLYPIVSS NKDYVHIRNV LRTMVVTILQ KVEGNQLVLR KPMTGQQYA IKNTGPFPWE KGDTLTLIPP LSTHSEEKLL KLGDWELTVP LVVPTAIAAE INIRLLCIGL IAVHREYNEM Q TIIDELCS ...String: METVYCTFDH KLSLSDISTL CKLMNIVIPI PAHHHLIGSG NLGLYPIVSS NKDYVHIRNV LRTMVVTILQ KVEGNQLVLR KPMTGQQYA IKNTGPFPWE KGDTLTLIPP LSTHSEEKLL KLGDWELTVP LVVPTAIAAE INIRLLCIGL IAVHREYNEM Q TIIDELCS IQYRDVLIKL PDIVNDKQSM YSMKTACISL SMITAMAPDI VRTYIDRLTL EDHSMLLIKC QELLSKRTTL ST QRCGQLH ATDIKDELKK IKSVLTMIDQ INSLTNEKTY FVVCDVSADN RMATCIYKN UniProtKB: Triplex capsid protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS buffer, pH 7.4 |
|---|---|
| Grid | Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: The grids were manually plunged into the ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 4828 / Average electron dose: 23.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.2 µm / Calibrated defocus min: 2.2 µm / Calibrated magnification: 64000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 2.2 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-6q1f: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)