+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2048 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the UPF-EJC complex | |||||||||
 Map data Map data | Cryo-EM reconstruction of the nonsense-mediated mRNA decay (NMD) complex containing proteins UPF1, UPF2, UPF3 and the Exon Junction Complex (EJC). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NMD / EJC / UPF1 / UPF2 / UPF3 / RNA degradation / GraFix | |||||||||
| Function / homology | WIBG, Mago-binding / UPF3 domain / Btz domain / RNA helicase UPF1, Cys/His rich zinc-binding domain / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Melero R / Buchwald G / Castano R / Raabe M / Gil D / Lazaro M / Urlaub H / Conti E / Llorca O | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2012 Journal: Nat Struct Mol Biol / Year: 2012Title: The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3' end. Authors: Roberto Melero / Gretel Buchwald / Raquel Castaño / Monika Raabe / David Gil / Melisa Lázaro / Henning Urlaub / Elena Conti / Oscar Llorca /  Abstract: Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance pathway that degrades aberrant mRNAs containing premature termination codons (PTCs). NMD is triggered upon the assembly of the UPF ...Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance pathway that degrades aberrant mRNAs containing premature termination codons (PTCs). NMD is triggered upon the assembly of the UPF surveillance complex near a PTC. In humans, UPF assembly is prompted by the exon junction complex (EJC). We investigated the molecular architecture of the human UPF complex bound to the EJC by cryo-EM and using positional restraints from additional EM, MS and biochemical interaction data. The heptameric assembly is built around UPF2, a scaffold protein with a ring structure that closes around the CH domain of UPF1, keeping the helicase region in an accessible and unwinding-competent state. UPF2 also positions UPF3 to interact with the EJC. The geometry is such that this transient complex poises UPF1 to elicit helicase activity toward the 3' end of the mRNP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2048.map.gz emd_2048.map.gz | 6.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2048-v30.xml emd-2048-v30.xml emd-2048.xml emd-2048.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2048.tif EMD-2048.tif | 1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2048 http://ftp.pdbj.org/pub/emdb/structures/EMD-2048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2048 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2048.map.gz / Format: CCP4 / Size: 9.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2048.map.gz / Format: CCP4 / Size: 9.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the nonsense-mediated mRNA decay (NMD) complex containing proteins UPF1, UPF2, UPF3 and the Exon Junction Complex (EJC). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
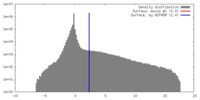
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex containing the NMD proteins UPF1, UPF2 and UPF3 bound to ...
| Entire | Name: Complex containing the NMD proteins UPF1, UPF2 and UPF3 bound to the exon junction complex (EJC) |
|---|---|
| Components |
|
-Supramolecule #1000: Complex containing the NMD proteins UPF1, UPF2 and UPF3 bound to ...
| Supramolecule | Name: Complex containing the NMD proteins UPF1, UPF2 and UPF3 bound to the exon junction complex (EJC) type: sample / ID: 1000 / Oligomeric state: Hetero-octameric / Number unique components: 8 |
|---|---|
| Molecular weight | Experimental: 390 KDa / Theoretical: 390 KDa |
-Macromolecule #1: Up-frameshift protein 1
| Macromolecule | Name: Up-frameshift protein 1 / type: protein_or_peptide / ID: 1 / Name.synonym: UPF1 / Details: 115-914 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 90 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: RNA helicase UPF1, Cys/His rich zinc-binding domain |
-Macromolecule #2: Up-frameshift protein 2
| Macromolecule | Name: Up-frameshift protein 2 / type: protein_or_peptide / ID: 2 / Name.synonym: UPF2 / Details: full length / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 148 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: RNA helicase UPF1, Cys/His rich zinc-binding domain |
-Macromolecule #3: Up-frameshift protein 3b
| Macromolecule | Name: Up-frameshift protein 3b / type: protein_or_peptide / ID: 3 / Name.synonym: UPF3b / Details: full length / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 56 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: UPF3 domain |
-Macromolecule #4: MAGO
| Macromolecule | Name: MAGO / type: protein_or_peptide / ID: 4 / Name.synonym: MAGO / Details: full length / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 16 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: WIBG, Mago-binding |
-Macromolecule #5: Y14
| Macromolecule | Name: Y14 / type: protein_or_peptide / ID: 5 / Name.synonym: Y14 / Details: residues 66-154 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 10 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: WIBG, Mago-binding |
-Macromolecule #6: Btz
| Macromolecule | Name: Btz / type: protein_or_peptide / ID: 6 / Name.synonym: Btz / Details: residues 136-286 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 17 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: Btz domain |
-Macromolecule #7: eIF4AIII
| Macromolecule | Name: eIF4AIII / type: protein_or_peptide / ID: 7 / Name.synonym: eIF4AIII / Details: full length / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 45 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay InterPro: Btz domain |
-Macromolecule #8: (U)8 RNA
| Macromolecule | Name: (U)8 RNA / type: rna / ID: 8 / Name.synonym: RNA / Details: Oligo(U) 8nt / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 3 KDa |
| Sequence | String: UUUUUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.175 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM K-phosphate,150 mM NaCl, 3 mM MgCl2, 20% sucrose, 0.1% glutaraldehyde |
| Grid | Details: Quantifoil grids (R2/2) with thin carbon film on top |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK III Method: Manual Application (3.5 microliters) Humidifier Off During Process Blot offset: -2 mm Blot Total: 2 Blot Time: 2 s Wait Time: 30 s Drain Time: 1 s |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Temperature | Min: 83 K / Max: 100 K / Average: 91.5 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected calculating the Fourier Transform of the CCD frames. Phase flipping |
| Specialist optics | Energy filter - Name: FEI / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 10.0 eV |
| Date | Oct 20, 2010 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Average electron dose: 15 e/Å2 / Details: 0.22 nm per pixel, final sampling / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 69494 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.112 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric. 626 cryo-holder (Gatan Inc., Warrendale, PA, USA) Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each CCD Frame using BSOFT |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, XMIPP, SPIDER, BSOFT Details: Initial reference obtained by random conical tilt of negative-stained images Number images used: 85000 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: R-factor |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: R-factor |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: R-factor |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)