[English] 日本語
 Yorodumi
Yorodumi- EMDB-20462: Escherichia coli RNA polymerase promoter unwinding intermediate (... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20462 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5a) with TraR and mutant rpsT P2 promoter | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRANSCRIPTION / transcription-dna complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsigma factor antagonist complex / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility ...sigma factor antagonist complex / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / DNA-directed RNA polymerase complex / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / protein dimerization activity / response to antibiotic / negative regulation of DNA-templated transcription / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / metal ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
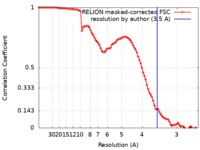
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Chen J / Chiu CE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Stepwise Promoter Melting by Bacterial RNA Polymerase. Authors: James Chen / Courtney Chiu / Saumya Gopalkrishnan / Albert Y Chen / Paul Dominic B Olinares / Ruth M Saecker / Jared T Winkelman / Michael F Maloney / Brian T Chait / Wilma Ross / Richard L ...Authors: James Chen / Courtney Chiu / Saumya Gopalkrishnan / Albert Y Chen / Paul Dominic B Olinares / Ruth M Saecker / Jared T Winkelman / Michael F Maloney / Brian T Chait / Wilma Ross / Richard L Gourse / Elizabeth A Campbell / Seth A Darst /  Abstract: Transcription initiation requires formation of the open promoter complex (RPo). To generate RPo, RNA polymerase (RNAP) unwinds the DNA duplex to form the transcription bubble and loads the DNA into ...Transcription initiation requires formation of the open promoter complex (RPo). To generate RPo, RNA polymerase (RNAP) unwinds the DNA duplex to form the transcription bubble and loads the DNA into the RNAP active site. RPo formation is a multi-step process with transient intermediates of unknown structure. We use single-particle cryoelectron microscopy to visualize seven intermediates containing Escherichia coli RNAP with the transcription factor TraR en route to forming RPo. The structures span the RPo formation pathway from initial recognition of the duplex promoter in a closed complex to the final RPo. The structures and supporting biochemical data define RNAP and promoter DNA conformational changes that delineate steps on the pathway, including previously undetected transient promoter-RNAP interactions that contribute to populating the intermediates but do not occur in RPo. Our work provides a structural basis for understanding RPo formation and its regulation, a major checkpoint in gene expression throughout evolution. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20462.map.gz emd_20462.map.gz | 60 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20462-v30.xml emd-20462-v30.xml emd-20462.xml emd-20462.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20462_fsc.xml emd_20462_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20462.png emd_20462.png | 47.1 KB | ||
| Filedesc metadata |  emd-20462.cif.gz emd-20462.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20462 http://ftp.pdbj.org/pub/emdb/structures/EMD-20462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20462 | HTTPS FTP |
-Validation report
| Summary document |  emd_20462_validation.pdf.gz emd_20462_validation.pdf.gz | 673.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20462_full_validation.pdf.gz emd_20462_full_validation.pdf.gz | 673 KB | Display | |
| Data in XML |  emd_20462_validation.xml.gz emd_20462_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_20462_validation.cif.gz emd_20462_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20462 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20462 | HTTPS FTP |
-Related structure data
| Related structure data |  6pssMC  6psqC  6psrC  6pstC  6psuC  6psvC  6pswC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20462.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20462.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Escherichia coli sigma70-holoenzyme bound to TraR and mutant rpsT...
+Supramolecule #1: Escherichia coli sigma70-holoenzyme bound to TraR and mutant rpsT...
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: RNA polymerase sigma factor RpoD
+Macromolecule #6: Protein TraR
+Macromolecule #7: DNA (85-MER)
+Macromolecule #8: DNA (85-MER)
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)