+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20350 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VC-Tn6677 multisubunit CRISPR/Cas effector, open conformation. | |||||||||
 Map data Map data | post-processed map using B-factor automatically calculated by Relion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR/Cas / Cascade / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Biological species |  | |||||||||
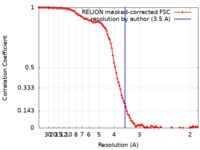
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Halpin-Healy T / Klompe S | |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Authors: Tyler S Halpin-Healy / Sanne E Klompe / Samuel H Sternberg / Israel S Fernández /  Abstract: Bacteria use adaptive immune systems encoded by CRISPR and Cas genes to maintain genomic integrity when challenged by pathogens and mobile genetic elements. Type I CRISPR-Cas systems typically ...Bacteria use adaptive immune systems encoded by CRISPR and Cas genes to maintain genomic integrity when challenged by pathogens and mobile genetic elements. Type I CRISPR-Cas systems typically target foreign DNA for degradation via joint action of the ribonucleoprotein complex Cascade and the helicase-nuclease Cas3, but nuclease-deficient type I systems lacking Cas3 have been repurposed for RNA-guided transposition by bacterial Tn7-like transposons. How CRISPR- and transposon-associated machineries collaborate during DNA targeting and insertion remains unknown. Here we describe structures of a TniQ-Cascade complex encoded by the Vibrio cholerae Tn6677 transposon using cryo-electron microscopy, revealing the mechanistic basis of this functional coupling. The cryo-electron microscopy maps enabled de novo modelling and refinement of the transposition protein TniQ, which binds to the Cascade complex as a dimer in a head-to-tail configuration, at the interface formed by Cas6 and Cas7 near the 3' end of the CRISPR RNA (crRNA). The natural Cas8-Cas5 fusion protein binds the 5' crRNA handle and contacts the TniQ dimer via a flexible insertion domain. A target DNA-bound structure reveals critical interactions necessary for protospacer-adjacent motif recognition and R-loop formation. This work lays the foundation for a structural understanding of how DNA targeting by TniQ-Cascade leads to downstream recruitment of additional transposase proteins, and will guide protein engineering efforts to leverage this system for programmable DNA insertions in genome-engineering applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20350.map.gz emd_20350.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20350-v30.xml emd-20350-v30.xml emd-20350.xml emd-20350.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20350_fsc.xml emd_20350_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_20350.png emd_20350.png | 177.4 KB | ||
| Masks |  emd_20350_msk_1.map emd_20350_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20350.cif.gz emd-20350.cif.gz | 6.7 KB | ||
| Others |  emd_20350_additional_1.map.gz emd_20350_additional_1.map.gz emd_20350_additional_2.map.gz emd_20350_additional_2.map.gz emd_20350_half_map_1.map.gz emd_20350_half_map_1.map.gz emd_20350_half_map_2.map.gz emd_20350_half_map_2.map.gz | 10.9 MB 80.8 MB 81.1 MB 81.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20350 http://ftp.pdbj.org/pub/emdb/structures/EMD-20350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20350 | HTTPS FTP |
-Validation report
| Summary document |  emd_20350_validation.pdf.gz emd_20350_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20350_full_validation.pdf.gz emd_20350_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_20350_validation.xml.gz emd_20350_validation.xml.gz | 17.3 KB | Display | |
| Data in CIF |  emd_20350_validation.cif.gz emd_20350_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20350 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20350 | HTTPS FTP |
-Related structure data
| Related structure data |  6pigMC  6pifC  6pijC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20350.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20350.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed map using B-factor automatically calculated by Relion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20350_msk_1.map emd_20350_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: masked post-processed map using B-factor automatically calculated by...
| File | emd_20350_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | masked post-processed map using B-factor automatically calculated by Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_20350_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1 from Relion3
| File | emd_20350_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 from Relion3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2 from Relion3
| File | emd_20350_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 from Relion3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VC-Tn667 transposon encoded CRISPR/Cas effector
| Entire | Name: VC-Tn667 transposon encoded CRISPR/Cas effector |
|---|---|
| Components |
|
-Supramolecule #1: VC-Tn667 transposon encoded CRISPR/Cas effector
| Supramolecule | Name: VC-Tn667 transposon encoded CRISPR/Cas effector / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: RNA (60-MER)
| Macromolecule | Name: RNA (60-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.237338 KDa |
| Sequence | String: CUGAUAACUU ACAGGACGCU UUGGCUUCAU UGCUUUUCAG GUGAACUGCC GAGUAGGUAG |
-Macromolecule #2: cas7 type I-F CRISPR-associated protein Csy3
| Macromolecule | Name: cas7 type I-F CRISPR-associated protein Csy3 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.020035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KLPTNLAYER SIDPSDVCFF VVWPDDRKTP LTYNSRTLLG QMEAKSLAYD VSGQPIKSAT AEALAQGNPH QVDFCHVPYG ASHIECSFS VSFSSELRQP YKCNSSKVKQ TLVQLVELYE TKIGWTELAT RYLMNICNGK WLWKNTRKAY CWNIVLTPWP W NGEKVGFE ...String: KLPTNLAYER SIDPSDVCFF VVWPDDRKTP LTYNSRTLLG QMEAKSLAYD VSGQPIKSAT AEALAQGNPH QVDFCHVPYG ASHIECSFS VSFSSELRQP YKCNSSKVKQ TLVQLVELYE TKIGWTELAT RYLMNICNGK WLWKNTRKAY CWNIVLTPWP W NGEKVGFE DIRTNYTSRQ DFKNNKNWSA IVEMIKTAFS STDGLAIFEV RATLHLPTNA MVRPSQVFTE KETQNSRVFQ ST TIDGERS PILGAFKTGA AIATIDDWYP EATEPLRVGR FGVHREDVTC YRHPSTGKDF FSILQQAEHY IEVLSANKTP AQE TINDMH FLMANLIKGG MFQHKGD |
-Macromolecule #3: cas5_8 naturally occurring fusion protein from Vibrio cholerae tr...
| Macromolecule | Name: cas5_8 naturally occurring fusion protein from Vibrio cholerae transposon Tn6677 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.064711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LKEIIASNPD DLTTELKRAF RPLTPHIAID GNEIDALTIL VNLTDKAKCK QKLRDEKWWA SCINCVNYRQ SHNPKFPDIR SEGVIRTQA LGELPSFLLS SSKIPPYHWS YSHDSKYVNK SAFLTNEFCW DGEISCLGEL LKDADHPLWN TLKKLGCSQK T CKAMAKQL ...String: LKEIIASNPD DLTTELKRAF RPLTPHIAID GNEIDALTIL VNLTDKAKCK QKLRDEKWWA SCINCVNYRQ SHNPKFPDIR SEGVIRTQA LGELPSFLLS SSKIPPYHWS YSHDSKYVNK SAFLTNEFCW DGEISCLGEL LKDADHPLWN TLKKLGCSQK T CKAMAKQL ADITLTTINV TLAPNYLTQI SLPDSDTSYI SLSPVASLSM QSHFHQRLQD ENRHSAITRF SRTTNMGVTA MT CGGAFRM LKSGAKFSSP PHHRLNNGSF LVLPNIRVCG ATALSSPVTV GIPSLTAFFG FVHAFERNIN RTTSSFRVES FAI CVHQLH VEKRGLTAEF VEKGDGTISA PATRDDWQCD VVFSLILNTN FAQHIDQDTL VTSLPKRLAR GSAKIAIDDF KHIN SFSTL ETAIESLPIE AGRWLSLYAQ SNNNLSDLLA AMTEDHQLMA SCVGYHLLEE PKDKPNSLRG YKHAIAECII GLINS ITFS SETDPNTIFW SLKNYQNYLV VQPRSIN |
-Macromolecule #4: type I-F CRISPR-associated endoribonuclease Cas6/Csy4
| Macromolecule | Name: type I-F CRISPR-associated endoribonuclease Cas6/Csy4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.926184 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KWYYKTITFL PELCNNESLA AKCLRVLHGF NYQYETRNIG VSFPLWCDAT VGKKISFVSK NKIELDLLLK QHYFVQMEQL QYFHISNTV LVPEDCTYVS FRRCQSIDKL TAAGLARKIR RLEKRALSRG EAFDPSSFAQ KEHTAIAHYH SLGESSKQTN R NFRLNIRM ...String: KWYYKTITFL PELCNNESLA AKCLRVLHGF NYQYETRNIG VSFPLWCDAT VGKKISFVSK NKIELDLLLK QHYFVQMEQL QYFHISNTV LVPEDCTYVS FRRCQSIDKL TAAGLARKIR RLEKRALSRG EAFDPSSFAQ KEHTAIAHYH SLGESSKQTN R NFRLNIRM LSEQPREGNS IFSSYGLANS ENSFQPVPLI |
-Macromolecule #5: TniQ monomer 1
| Macromolecule | Name: TniQ monomer 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.51716 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AMFLQRPKPY SDESLESFFI RVANKNGYGD VHRFLEATKR FLQDIDHNGY QTFPTDITRI NPYSAKNSSS ARTASFLKLA QLTFNEPPE LLGLAINRTN MKYSPSTSAV VRGAEVFPRS LLRTHSIPCC PLCLRENGYA SYLWHFQGYE YCHSHNVPLI T TCSGHEAA ...String: AMFLQRPKPY SDESLESFFI RVANKNGYGD VHRFLEATKR FLQDIDHNGY QTFPTDITRI NPYSAKNSSS ARTASFLKLA QLTFNEPPE LLGLAINRTN MKYSPSTSAV VRGAEVFPRS LLRTHSIPCC PLCLRENGYA SYLWHFQGYE YCHSHNVPLI T TCSGHEAA CTVSNWLAGH ESKPLPNLPK SYRWGLVHWW MGIKDSDHFS FVQFFSNWPR SFHSIIEDEV EFNLEHAVVS TS ELRLKDL LGRLFFGSIR LPERNLQHNI ILGELLCYLE NRLWQDKGLI ANLKMNALEA TVMLNCSLDQ IASMVEQRIL KPN RKSKDV TDYLFHFGDI FCLWLAEFQS DEFNRSFYVS RW |
-Macromolecule #6: TniQ monomer 2
| Macromolecule | Name: TniQ monomer 2 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.315031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AMFLQRPKPY SDESLESFFI RVANKNGYGD VHRFLEATKR FLQDIDHNGY QTFPTDITRI NPYSAKNSSS ARTASFLKLA QLTFNEPPE LLGLAINRTN MKYSPSTSAV VRGAEVFPRS LLRTHSIPCC PLCLRENGYA SYLWHFQGYE YCHSHNVPLI T TCSCGKEF ...String: AMFLQRPKPY SDESLESFFI RVANKNGYGD VHRFLEATKR FLQDIDHNGY QTFPTDITRI NPYSAKNSSS ARTASFLKLA QLTFNEPPE LLGLAINRTN MKYSPSTSAV VRGAEVFPRS LLRTHSIPCC PLCLRENGYA SYLWHFQGYE YCHSHNVPLI T TCSCGKEF DYRVSEAACT VSNWLAGHES KPLPNLPKSY RWGLVHWWMG IKDDHFSFVQ FFSNWPRSFH SIIEDEVEFN LE HAVVSTS ELRLKDLLGR LFFGSIRLPE RNLQHNIILG ELLCYLENRL WQDKGLIANL KMNALEATVM LNCSLDQIAS MVE QRILKP NAAAAAAAAA DVTDYLFHFG DIFCLWLAAF QSDEFNRSFY VSR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)