[English] 日本語
 Yorodumi
Yorodumi- EMDB-20265: Cryo-EM structure of voltage-gated sodium channel NavAb N49K/L109... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20265 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of voltage-gated sodium channel NavAb N49K/L109A/M116V/G94C/Q150C disulfide crosslinked mutant in the resting state | |||||||||||||||
 Map data Map data | Cryo-EM map of disulfide crosslinked MBP-NavAb N49K/L109A/M116V/G9C/Q150C in the resting state | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ion channel / ion transport protein / MEMBRANE PROTEIN / metal transport | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated sodium channel complex / voltage-gated sodium channel activity / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing ...voltage-gated sodium channel complex / voltage-gated sodium channel activity / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space / periplasmic space / DNA damage response / metal ion binding / identical protein binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Arcobacter butzleri RM4018 (bacteria) / Arcobacter butzleri RM4018 (bacteria) /  Arcobacter butzleri (strain RM4018) (bacteria) Arcobacter butzleri (strain RM4018) (bacteria) | |||||||||||||||
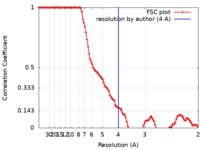
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Wisedchaisri G / Tonggu L | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel. Authors: Goragot Wisedchaisri / Lige Tonggu / Eedann McCord / Tamer M Gamal El-Din / Liguo Wang / Ning Zheng / William A Catterall /  Abstract: Voltage-gated sodium (Na) channels initiate action potentials in nerve, muscle, and other electrically excitable cells. The structural basis of voltage gating is uncertain because the resting state ...Voltage-gated sodium (Na) channels initiate action potentials in nerve, muscle, and other electrically excitable cells. The structural basis of voltage gating is uncertain because the resting state exists only at deeply negative membrane potentials. To stabilize the resting conformation, we inserted voltage-shifting mutations and introduced a disulfide crosslink in the VS of the ancestral bacterial sodium channel NaAb. Here, we present a cryo-EM structure of the resting state and a complete voltage-dependent gating mechanism. The S4 segment of the VS is drawn intracellularly, with three gating charges passing through the transmembrane electric field. This movement forms an elbow connecting S4 to the S4-S5 linker, tightens the collar around the S6 activation gate, and prevents its opening. Our structure supports the classical "sliding helix" mechanism of voltage sensing and provides a complete gating mechanism for voltage sensor function, pore opening, and activation-gate closure based on high-resolution structures of a single sodium channel protein. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20265.map.gz emd_20265.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20265-v30.xml emd-20265-v30.xml emd-20265.xml emd-20265.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20265_fsc.xml emd_20265_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_20265.png emd_20265.png | 162.1 KB | ||
| Filedesc metadata |  emd-20265.cif.gz emd-20265.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20265 http://ftp.pdbj.org/pub/emdb/structures/EMD-20265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20265 | HTTPS FTP |
-Validation report
| Summary document |  emd_20265_validation.pdf.gz emd_20265_validation.pdf.gz | 351.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20265_full_validation.pdf.gz emd_20265_full_validation.pdf.gz | 351.3 KB | Display | |
| Data in XML |  emd_20265_validation.xml.gz emd_20265_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_20265_validation.cif.gz emd_20265_validation.cif.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20265 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20265 | HTTPS FTP |
-Related structure data
| Related structure data |  6p6wMC  6p6xC  6p6yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20265.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20265.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of disulfide crosslinked MBP-NavAb N49K/L109A/M116V/G9C/Q150C in the resting state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.056 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fusion of maltose-binding protein and voltage-gated sodium channe...
| Entire | Name: Fusion of maltose-binding protein and voltage-gated sodium channel NavAb in the resting state |
|---|---|
| Components |
|
-Supramolecule #1: Fusion of maltose-binding protein and voltage-gated sodium channe...
| Supramolecule | Name: Fusion of maltose-binding protein and voltage-gated sodium channel NavAb in the resting state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Arcobacter butzleri RM4018 (bacteria) Arcobacter butzleri RM4018 (bacteria) |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Fusion of Maltose-binding protein and voltage-gated sodium channe...
| Macromolecule | Name: Fusion of Maltose-binding protein and voltage-gated sodium channel NavAb type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Arcobacter butzleri (strain RM4018) (bacteria) / Strain: RM4018 Arcobacter butzleri (strain RM4018) (bacteria) / Strain: RM4018 |
| Molecular weight | Theoretical: 74.072797 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKIKTGARIL ALSALTTMMF SASALAKIEE GKLVIWINGD KGYNGLAEVG KKFEKDTGIK VTVEHPDKLE EKFPQVAATG DGPDIIFWA HDRFGGYAQS GLLAEITPDK AFQDKLYPFT WDAVRYNGKL IAYPIAVEAL SLIYNKDLLP NPPKTWEEIP A LDKELKAK ...String: MKIKTGARIL ALSALTTMMF SASALAKIEE GKLVIWINGD KGYNGLAEVG KKFEKDTGIK VTVEHPDKLE EKFPQVAATG DGPDIIFWA HDRFGGYAQS GLLAEITPDK AFQDKLYPFT WDAVRYNGKL IAYPIAVEAL SLIYNKDLLP NPPKTWEEIP A LDKELKAK GKSALMFNLQ EPYFTWPLIA ADGGYAFKYE NGKYDIKDVG VDNAGAKAGL TFLVDLIKNK HMNADTDYSI AE AAFNKGE TAMTINGPWA WSNIDTSKVN YGVTVLPTFK GQPSKPFVGV LSAGINAASP NKELAKEFLE NYLLTDEGLE AVN KDKPLG AVALKSYEEE LAKDPRIAAT MENAQKGEIM PNIPQMSAFW YAVRTAVINA ASGRQTVDEA LKDAQTNAMY LAIT NIVES SFFTKFIIYL IVLNGITMGL ETSKTFMQSF GVYTTLFKQI VITIFTIEII LRIYVHRISF FKDPWSLFDF FVVAI SLVP TSSCFEILRV LRVLRLFRAV TAVPQVRKIV SALISVIPGM LSVIALMTLF FYIFAIMATC LFGERFPEWF GTLGES FYT LFQVMTLESW SMGIVRPLME VYPYAWVFFI PFIFVVTFVM INLVVAIIVD AMAILNQKEE QHIIDEVQSH EDNINNE II KLREEIVELK ELIKTSLKN UniProtKB: Maltodextrin-binding protein, Ion transport protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.04 kPa | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 40 % / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90.8 K / Max: 103.5 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-43 / Number grids imaged: 9 / Number real images: 5000 / Average exposure time: 8.6 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 1001-1221 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-6p6w: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)