+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-2023 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
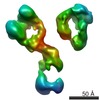
| タイトル | Three-dimensional reconstruction of a targeted individual 17nm nascent high-density lipoprotein (HDL) particle by individual-particle electron tomography (IPET). The reconstruction displayed a ring-shaped structure of containing three protein - apoipoprotein A-Is (28 kDa each). | |||||||||
 マップデータ マップデータ | 17nm Nascent HDL particle Number 1 by cryo-ET | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | reconstituted 17nm nascent high-density lipoprotein (HDl) | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | サブトモグラム平均法 / クライオ電子顕微鏡法 / ネガティブ染色法 / 解像度: 42.0 Å | |||||||||
 データ登録者 データ登録者 | Zhang L / Ren G | |||||||||
 引用 引用 |  ジャーナル: PLoS One / 年: 2012 ジャーナル: PLoS One / 年: 2012タイトル: IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. 著者: Lei Zhang / Gang Ren /  要旨: The dynamic personalities and structural heterogeneity of proteins are essential for proper functioning. Structural determination of dynamic/heterogeneous proteins is limited by conventional ...The dynamic personalities and structural heterogeneity of proteins are essential for proper functioning. Structural determination of dynamic/heterogeneous proteins is limited by conventional approaches of X-ray and electron microscopy (EM) of single-particle reconstruction that require an average from thousands to millions different molecules. Cryo-electron tomography (cryoET) is an approach to determine three-dimensional (3D) reconstruction of a single and unique biological object such as bacteria and cells, by imaging the object from a series of tilting angles. However, cconventional reconstruction methods use large-size whole-micrographs that are limited by reconstruction resolution (lower than 20 Å), especially for small and low-symmetric molecule (<400 kDa). In this study, we demonstrated the adverse effects from image distortion and the measuring tilt-errors (including tilt-axis and tilt-angle errors) both play a major role in limiting the reconstruction resolution. Therefore, we developed a "focused electron tomography reconstruction" (FETR) algorithm to improve the resolution by decreasing the reconstructing image size so that it contains only a single-instance protein. FETR can tolerate certain levels of image-distortion and measuring tilt-errors, and can also precisely determine the translational parameters via an iterative refinement process that contains a series of automatically generated dynamic filters and masks. To describe this method, a set of simulated cryoET images was employed; to validate this approach, the real experimental images from negative-staining and cryoET were used. Since this approach can obtain the structure of a single-instance molecule/particle, we named it individual-particle electron tomography (IPET) as a new robust strategy/approach that does not require a pre-given initial model, class averaging of multiple molecules or an extended ordered lattice, but can tolerate small tilt-errors for high-resolution single "snapshot" molecule structure determination. Thus, FETR/IPET provides a completely new opportunity for a single-molecule structure determination, and could be used to study the dynamic character and equilibrium fluctuation of macromolecules. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_2023.map.gz emd_2023.map.gz | 2.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-2023-v30.xml emd-2023-v30.xml emd-2023.xml emd-2023.xml | 10.7 KB 10.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  EMD-2023.tif EMD-2023.tif | 65.1 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2023 http://ftp.pdbj.org/pub/emdb/structures/EMD-2023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2023 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_2023_validation.pdf.gz emd_2023_validation.pdf.gz | 194.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_2023_full_validation.pdf.gz emd_2023_full_validation.pdf.gz | 193.5 KB | 表示 | |
| XML形式データ |  emd_2023_validation.xml.gz emd_2023_validation.xml.gz | 5.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2023 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2023 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_2023.map.gz / 形式: CCP4 / 大きさ: 3.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_2023.map.gz / 形式: CCP4 / 大きさ: 3.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | 17nm Nascent HDL particle Number 1 by cryo-ET | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 3.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : a 17nm nascent HDL particle
| 全体 | 名称: a 17nm nascent HDL particle |
|---|---|
| 要素 |
|
-超分子 #1000: a 17nm nascent HDL particle
| 超分子 | 名称: a 17nm nascent HDL particle / タイプ: sample / ID: 1000 / 詳細: fresh sample was prepared with 2 days before using. / 集合状態: monomer / Number unique components: 1 |
|---|---|
| 分子量 | 実験値: 200 KDa / 理論値: 200 KDa |
-分子 #1: 17 nm nascent HDL
| 分子 | 名称: 17 nm nascent HDL / タイプ: protein_or_peptide / ID: 1 / Name.synonym: 17 nm nascent HDL 詳細: HDL contains lipids and three apoA-I molecules. Each A-I molecular weight is about 28kDa. コピー数: 1 / 集合状態: monomer / 組換発現: No / データベース: NCBI |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) / 株: nascent HDL / 別称: Human / 細胞中の位置: Plasma Homo sapiens (ヒト) / 株: nascent HDL / 別称: Human / 細胞中の位置: Plasma |
| 分子量 | 実験値: 200 KDa / 理論値: 200 KDa |
-実験情報
-構造解析
| 手法 | ネガティブ染色法, クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | サブトモグラム平均法 |
- 試料調製
試料調製
| 濃度 | 0.01 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 詳細: 1X Dulbeccos phosphate-buffered saline (Invitrogen, La Jolla, CA), 2.7 mM KCl, 1.46 mM KH2PO4, 136.9 mM NaCl, and 8.1 mM Na2HPO4 |
| 染色 | タイプ: NEGATIVE 詳細: EM Specimens were prepared by standard cryo-EM protocol. |
| グリッド | 詳細: 200 mesh glow-discharged holey thin carbon-coated EM grids (Cu-200HN, Pacific Grid-Tech, USA) |
| 凍結 | 凍結剤: NITROGEN / チャンバー内湿度: 90 % / チャンバー内温度: 103 K / 装置: HOMEMADE PLUNGER / 詳細: Vitrification instrument: manual / 手法: Blot for 2 seconds before plunging |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI 12 |
|---|---|
| 温度 | 平均: 100 K |
| 詳細 | Tilt step is 1.5 degree |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: GATAN ULTRASCAN 4000 (4k x 4k) デジタル化 - サンプリング間隔: 1.73 µm / 実像数: 81 / 平均電子線量: 140 e/Å2 / ビット/ピクセル: 16 |
| 電子線 | 加速電圧: 120 kV / 電子線源: LAB6 |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.2 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 67000 |
| 試料ステージ | 試料ホルダー: Gatan / 試料ホルダーモデル: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
- 画像解析
画像解析
| 詳細 | Single targeted particle"s images were reconstructed by focus ET reconstruction (FETR) algorithm of individual particle electron tomography (IPET) method. Average number of tilts used in the 3D reconstructions: 81. Average tomographic tilt angle increment: 1.5. |
|---|---|
| 最終 再構成 | アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 42.0 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: IPET, and, FETR 詳細: Map was reconstructed by individual-particle electron tomography (IPET)and Focus ET Reconstruction Algorithm. |
| CTF補正 | 詳細: TOMOCTF |
| 最終 角度割当 | 詳細: Tomography tilt angle from -60 to 60 in step of 1.5 |
 ムービー
ムービー コントローラー
コントローラー



 UCSF Chimera
UCSF Chimera




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















