[English] 日本語
 Yorodumi
Yorodumi- EMDB-20092: Monomeric kinesin-1 motor domain in no-nucleotide state bound to ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20092 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Monomeric kinesin-1 motor domain in no-nucleotide state bound to GMPCPP-stabilized microtubule | |||||||||
 Map data Map data | Monomeric kinesin-1 motor domain in the no-nucleotide state bound to GMPCPP-stabilized microtubule | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinesin / Microtubule / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of modification of synapse structure, modulating synaptic transmission / plus-end-directed vesicle transport along microtubule / cytoplasm organization / cytolytic granule membrane / anterograde dendritic transport of neurotransmitter receptor complex / anterograde neuronal dense core vesicle transport / mitocytosis / retrograde neuronal dense core vesicle transport / anterograde axonal protein transport / ciliary rootlet ...regulation of modification of synapse structure, modulating synaptic transmission / plus-end-directed vesicle transport along microtubule / cytoplasm organization / cytolytic granule membrane / anterograde dendritic transport of neurotransmitter receptor complex / anterograde neuronal dense core vesicle transport / mitocytosis / retrograde neuronal dense core vesicle transport / anterograde axonal protein transport / ciliary rootlet / lysosome localization / positive regulation of potassium ion transport / plus-end-directed microtubule motor activity / vesicle transport along microtubule / RHO GTPases activate KTN1 / Kinesins / positive regulation of axon guidance / kinesin complex / microtubule motor activity / centrosome localization / mitochondrion transport along microtubule / COPI-dependent Golgi-to-ER retrograde traffic / microtubule-based movement / stress granule disassembly / natural killer cell mediated cytotoxicity / Insulin processing / synaptic vesicle transport / postsynaptic cytosol / microtubule-based process / cytoplasmic microtubule / phagocytic vesicle / axon cytoplasm / MHC class II antigen presentation / cellular response to interleukin-4 / dendrite cytoplasm / axon guidance / positive regulation of synaptic transmission, GABAergic / regulation of membrane potential / positive regulation of protein localization to plasma membrane / cellular response to type II interferon / structural constituent of cytoskeleton / centriolar satellite / microtubule cytoskeleton organization / neuron migration / Signaling by ALK fusions and activated point mutants / mitotic cell cycle / double-stranded RNA binding / microtubule cytoskeleton / nuclear membrane / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / vesicle / microtubule / cilium / cadherin binding / protein heterodimerization activity / GTPase activity / ubiquitin protein ligase binding / GTP binding / protein-containing complex binding / perinuclear region of cytoplasm / ATP hydrolysis activity / mitochondrion / ATP binding / metal ion binding / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.67 Å | |||||||||
 Authors Authors | Cha HK / Debs G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural Intermediates of the Dimeric Kinesin Stepping Cycle Revealed by Cryo-EM Authors: Cha HK / Debs G / Liu X / Liu D / Sindelar CV | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20092.map.gz emd_20092.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20092-v30.xml emd-20092-v30.xml emd-20092.xml emd-20092.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20092.png emd_20092.png | 157.1 KB | ||
| Filedesc metadata |  emd-20092.cif.gz emd-20092.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20092 http://ftp.pdbj.org/pub/emdb/structures/EMD-20092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20092 | HTTPS FTP |
-Validation report
| Summary document |  emd_20092_validation.pdf.gz emd_20092_validation.pdf.gz | 402.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20092_full_validation.pdf.gz emd_20092_full_validation.pdf.gz | 401.7 KB | Display | |
| Data in XML |  emd_20092_validation.xml.gz emd_20092_validation.xml.gz | 7.9 KB | Display | |
| Data in CIF |  emd_20092_validation.cif.gz emd_20092_validation.cif.gz | 9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20092 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20092 | HTTPS FTP |
-Related structure data
| Related structure data |  6ojqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20092.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20092.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Monomeric kinesin-1 motor domain in the no-nucleotide state bound to GMPCPP-stabilized microtubule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
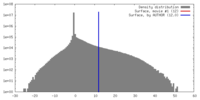
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Monomeric kinesin-1 motor domain in the no-nucleotide state bound...
| Entire | Name: Monomeric kinesin-1 motor domain in the no-nucleotide state bound to GMPCPP-stabilized microtubules |
|---|---|
| Components |
|
-Supramolecule #1: Monomeric kinesin-1 motor domain in the no-nucleotide state bound...
| Supramolecule | Name: Monomeric kinesin-1 motor domain in the no-nucleotide state bound to GMPCPP-stabilized microtubules type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin beta-2B chain
| Macromolecule | Name: Tubulin beta-2B chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.825859 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY Q UniProtKB: Tubulin beta-2B chain |
-Macromolecule #2: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.665027 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GV UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #3: Kinesin-1 heavy chain
| Macromolecule | Name: Kinesin-1 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.445801 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NIKVMCRFRP LNESEVNRGD KYIAKFQGED TVVIASKPYA FDRVFQSSTS QEQVYNDAAK KIVKDVLEGY NGTIFAYGQT SSGKTHTME GKLHDPEGMG IIPRIVQDIF NYIYSMDENL EFHIKVSYFE IYLDKIRDLL DVSKTNLSVH EDKNRVPYVK G ATERFVSS ...String: NIKVMCRFRP LNESEVNRGD KYIAKFQGED TVVIASKPYA FDRVFQSSTS QEQVYNDAAK KIVKDVLEGY NGTIFAYGQT SSGKTHTME GKLHDPEGMG IIPRIVQDIF NYIYSMDENL EFHIKVSYFE IYLDKIRDLL DVSKTNLSVH EDKNRVPYVK G ATERFVSS PDEVMDTIDE GKSNRHVAVT NMNEHSSRSH SIFLINVKQE NTQTEQKLSG KLYLVDLAGS EKVSKTGAEG AV LDEAKNI NKSLSALGNV ISALAEGSTY VPYRDSKMTR ILQDSLGGNA RTTIVICCSP SSYNESETKS TLLFGQRAKT UniProtKB: Kinesin-1 heavy chain |
-Macromolecule #4: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 4 / Number of copies: 1 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.625 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.67 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 403424 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)