[English] 日本語
 Yorodumi
Yorodumi- EMDB-19979: Inhibitor-free outward-open structure of Drosophila dopamine tran... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inhibitor-free outward-open structure of Drosophila dopamine transporter | ||||||||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | SLC6A3 / Dopamine transporter / neurotransmitter sodium symporters / MEMBRANE PROTEIN | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDopamine clearance from the synaptic cleft / SLC-mediated transport of neurotransmitters / circadian sleep/wake cycle / cocaine binding / response to odorant / norepinephrine transport / dopamine:sodium symporter activity / regulation of presynaptic cytosolic calcium ion concentration / dopamine transport / sleep ...Dopamine clearance from the synaptic cleft / SLC-mediated transport of neurotransmitters / circadian sleep/wake cycle / cocaine binding / response to odorant / norepinephrine transport / dopamine:sodium symporter activity / regulation of presynaptic cytosolic calcium ion concentration / dopamine transport / sleep / neuronal cell body membrane / dopamine uptake involved in synaptic transmission / amino acid transport / sodium ion transmembrane transport / adult locomotory behavior / presynaptic membrane / axon / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |   | ||||||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Pedersen CN / Yang F / Ita S / Xu Y / Akunuri R / Trampari S / Neumann CMT / Desdorf LM / Schioett B / Salvino JM ...Pedersen CN / Yang F / Ita S / Xu Y / Akunuri R / Trampari S / Neumann CMT / Desdorf LM / Schioett B / Salvino JM / Mortensen OV / Nissen P / Shahsavar A | ||||||||||||||||||||||||||||||||||||
| Funding support |  Denmark, Denmark,  United States, 11 items United States, 11 items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: J Neurochem / Year: 2024 Journal: J Neurochem / Year: 2024Title: Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site. Authors: Clara Nautrup Pedersen / Fuyu Yang / Samantha Ita / Yibin Xu / Ravikumar Akunuri / Sofia Trampari / Caroline Marie Teresa Neumann / Lasse Messell Desdorf / Birgit Schiøtt / Joseph M Salvino ...Authors: Clara Nautrup Pedersen / Fuyu Yang / Samantha Ita / Yibin Xu / Ravikumar Akunuri / Sofia Trampari / Caroline Marie Teresa Neumann / Lasse Messell Desdorf / Birgit Schiøtt / Joseph M Salvino / Ole Valente Mortensen / Poul Nissen / Azadeh Shahsavar /   Abstract: The regulation of dopamine (DA) removal from the synaptic cleft is a crucial process in neurotransmission and is facilitated by the sodium- and chloride-coupled dopamine transporter DAT. ...The regulation of dopamine (DA) removal from the synaptic cleft is a crucial process in neurotransmission and is facilitated by the sodium- and chloride-coupled dopamine transporter DAT. Psychostimulant drugs, cocaine, and amphetamine, both block the uptake of DA, while amphetamine also triggers the release of DA. As a result, they prolong or even amplify neurotransmitter signaling. Atypical inhibitors of DAT lack cocaine-like rewarding effects and offer a promising strategy for the treatment of drug use disorders. Here, we present the 3.2 Å resolution cryo-electron microscopy structure of the Drosophila melanogaster dopamine transporter (dDAT) in complex with the atypical non-competitive inhibitor AC-4-248. The inhibitor partially binds at the central binding site, extending into the extracellular vestibule, and locks the transporter in an outward open conformation. Our findings propose mechanisms for the non-competitive inhibition of DAT and attenuation of cocaine potency by AC-4-248 and provide a basis for the rational design of more efficacious atypical inhibitors. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19979.map.gz emd_19979.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19979-v30.xml emd-19979-v30.xml emd-19979.xml emd-19979.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19979.png emd_19979.png | 49.7 KB | ||
| Filedesc metadata |  emd-19979.cif.gz emd-19979.cif.gz | 7.3 KB | ||
| Others |  emd_19979_half_map_1.map.gz emd_19979_half_map_1.map.gz emd_19979_half_map_2.map.gz emd_19979_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19979 http://ftp.pdbj.org/pub/emdb/structures/EMD-19979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19979 | HTTPS FTP |
-Validation report
| Summary document |  emd_19979_validation.pdf.gz emd_19979_validation.pdf.gz | 795 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19979_full_validation.pdf.gz emd_19979_full_validation.pdf.gz | 794.6 KB | Display | |
| Data in XML |  emd_19979_validation.xml.gz emd_19979_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_19979_validation.cif.gz emd_19979_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19979 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19979 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19979 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19979 | HTTPS FTP |
-Related structure data
| Related structure data |  9eupMC  9euoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19979.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19979.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.294 Å | ||||||||||||||||||||||||||||||||||||
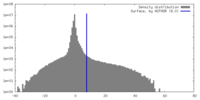
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_19979_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
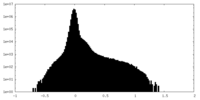
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_19979_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
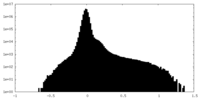
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Inhibitor-free Drosophila melanogaster dopamine transporter in co...
| Entire | Name: Inhibitor-free Drosophila melanogaster dopamine transporter in complex with Fab 9D5 |
|---|---|
| Components |
|
-Supramolecule #1: Inhibitor-free Drosophila melanogaster dopamine transporter in co...
| Supramolecule | Name: Inhibitor-free Drosophila melanogaster dopamine transporter in complex with Fab 9D5 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.602 KDa |
-Macromolecule #1: Sodium-dependent dopamine transporter
| Macromolecule | Name: Sodium-dependent dopamine transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.93952 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNSISDERET WSGKVDFLLS VIGFAVDLAN VWRFPYLCYK NGGGAFLVPY GIMLAVGGIP LFYMELALGQ HNRKGAITCW GRLVPLFKG IGYAVVLIAF YVDFYYNVII AWSLRFFFAS FTNSLPWTSC NNIWNTPNCR PFESQGFQSA ASEYFNRYIL E LNRSEGIH ...String: MNSISDERET WSGKVDFLLS VIGFAVDLAN VWRFPYLCYK NGGGAFLVPY GIMLAVGGIP LFYMELALGQ HNRKGAITCW GRLVPLFKG IGYAVVLIAF YVDFYYNVII AWSLRFFFAS FTNSLPWTSC NNIWNTPNCR PFESQGFQSA ASEYFNRYIL E LNRSEGIH DLGAIKWDMA LCLLIVYLIC YFSLWKGIST SGKVVWFTAL FPYAALLILL IRGLTLPGSF LGIQYYLTPN FS AIYKAEV WADAATQVFF SLGPGFGVLL AYASYNKYHN NVYKDALLTS FINSATSFIA GFVIFSVLGY MAHTLGVRIE DVA TEGPGL VFVVYPAAIA TMPASTFWAL IFFMMLATLG LDSSFGGSEA IITALSDEFP KIKRNRELFV AGLFSLYFVV GLAS CTQGG FYFFHLLDRY AAGYSILVAV FFEAIAVSWI YGTNRFSEDI RDMIGFPPGR YWQVCWRFVA PIFLLFITVY LLIGY EPLT YADYVYPSWA NALGWCIAGS SVVMIPAVAI FKLLSTPGSL RQRFTILTTP WRDQQLVPR UniProtKB: Sodium-dependent dopamine transporter |
-Macromolecule #2: 9D5 ANTIBODY, HEAVY CHAIN
| Macromolecule | Name: 9D5 ANTIBODY, HEAVY CHAIN / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.921338 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNFGLRLVFL VLILKGVQCE VQLVESGGGL VKPGGSLKLS CAASGFTFSS YAMSWVRQSP EKRLEWVAEI SSGGRYIYYS DTVTGRFTI SRDNARNILH LEMSSLRSED TAMYYCARGE VRQRGFDYWG QGTTLTVSSA KTTAPSVYPL APVCGDTTGS S VTLGCLVK ...String: MNFGLRLVFL VLILKGVQCE VQLVESGGGL VKPGGSLKLS CAASGFTFSS YAMSWVRQSP EKRLEWVAEI SSGGRYIYYS DTVTGRFTI SRDNARNILH LEMSSLRSED TAMYYCARGE VRQRGFDYWG QGTTLTVSSA KTTAPSVYPL APVCGDTTGS S VTLGCLVK GYFPEPVTLT WNSGSLSSGV HTFPAVLQSD LYTLSSSVTV TSSTWPSQSI TCNVAHPASS TKVDKKIEPR GP |
-Macromolecule #3: 9D5 ANTIBODY, LIGHT CHAIN
| Macromolecule | Name: 9D5 ANTIBODY, LIGHT CHAIN / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.840607 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDFQVQIFSF LLISASVAMS RGENVLTQSP AIMSTSPGEK VTMTCRASSS VGSSYLHWYQ QKSGASPKLW IYSTSNLASG VPARFSGSG SGTSYSLTIS SVEAEDAATY YCQQFSGYPL TFGSGTKLEM KRADAAPTVS IFPPSSEQLT SGGASVVCFL N NFYPKDIN ...String: MDFQVQIFSF LLISASVAMS RGENVLTQSP AIMSTSPGEK VTMTCRASSS VGSSYLHWYQ QKSGASPKLW IYSTSNLASG VPARFSGSG SGTSYSLTIS SVEAEDAATY YCQQFSGYPL TFGSGTKLEM KRADAAPTVS IFPPSSEQLT SGGASVVCFL N NFYPKDIN VKWKIDGSER QNGVLNSWTD QDSKDSTYSM SSTLTLTKDE YERHNSYTCE ATHKTSTSPI VKSFNRNEC |
-Macromolecule #4: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #5: TRIS-HYDROXYMETHYL-METHYL-AMMONIUM
| Macromolecule | Name: TRIS-HYDROXYMETHYL-METHYL-AMMONIUM / type: ligand / ID: 5 / Number of copies: 1 / Formula: 144 |
|---|---|
| Molecular weight | Theoretical: 122.143 Da |
| Chemical component information | 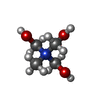 ChemComp-144: |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #7: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 7 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #8: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 4294 / Average electron dose: 59.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 137600 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)