[English] 日本語
 Yorodumi
Yorodumi- PDB-9eup: Inhibitor-free outward-open structure of Drosophila dopamine tran... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9eup | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inhibitor-free outward-open structure of Drosophila dopamine transporter | ||||||||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / SLC6A3 / Dopamine transporter / neurotransmitter sodium symporters | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information: / SLC-mediated transport of neurotransmitters / circadian sleep/wake cycle / response to odorant / cocaine binding / norepinephrine transport / dopamine:sodium symporter activity / regulation of presynaptic cytosolic calcium ion concentration / dopamine transport / sleep ...: / SLC-mediated transport of neurotransmitters / circadian sleep/wake cycle / response to odorant / cocaine binding / norepinephrine transport / dopamine:sodium symporter activity / regulation of presynaptic cytosolic calcium ion concentration / dopamine transport / sleep / neuronal cell body membrane / dopamine uptake involved in synaptic transmission / amino acid transport / sodium ion transmembrane transport / adult locomotory behavior / presynaptic membrane / axon / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |   | ||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Pedersen, C.N. / Yang, F. / Ita, S. / Xu, Y. / Akunuri, R. / Trampari, S. / Neumann, C.M.T. / Desdorf, L.M. / Schioett, B. / Salvino, J.M. ...Pedersen, C.N. / Yang, F. / Ita, S. / Xu, Y. / Akunuri, R. / Trampari, S. / Neumann, C.M.T. / Desdorf, L.M. / Schioett, B. / Salvino, J.M. / Mortensen, O.V. / Nissen, P. / Shahsavar, A. | ||||||||||||||||||||||||||||||||||||
| Funding support |  Denmark, Denmark,  United States, 11items United States, 11items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: J Neurochem / Year: 2024 Journal: J Neurochem / Year: 2024Title: Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site. Authors: Clara Nautrup Pedersen / Fuyu Yang / Samantha Ita / Yibin Xu / Ravikumar Akunuri / Sofia Trampari / Caroline Marie Teresa Neumann / Lasse Messell Desdorf / Birgit Schiøtt / Joseph M Salvino ...Authors: Clara Nautrup Pedersen / Fuyu Yang / Samantha Ita / Yibin Xu / Ravikumar Akunuri / Sofia Trampari / Caroline Marie Teresa Neumann / Lasse Messell Desdorf / Birgit Schiøtt / Joseph M Salvino / Ole Valente Mortensen / Poul Nissen / Azadeh Shahsavar /   Abstract: The regulation of dopamine (DA) removal from the synaptic cleft is a crucial process in neurotransmission and is facilitated by the sodium- and chloride-coupled dopamine transporter DAT. ...The regulation of dopamine (DA) removal from the synaptic cleft is a crucial process in neurotransmission and is facilitated by the sodium- and chloride-coupled dopamine transporter DAT. Psychostimulant drugs, cocaine, and amphetamine, both block the uptake of DA, while amphetamine also triggers the release of DA. As a result, they prolong or even amplify neurotransmitter signaling. Atypical inhibitors of DAT lack cocaine-like rewarding effects and offer a promising strategy for the treatment of drug use disorders. Here, we present the 3.2 Å resolution cryo-electron microscopy structure of the Drosophila melanogaster dopamine transporter (dDAT) in complex with the atypical non-competitive inhibitor AC-4-248. The inhibitor partially binds at the central binding site, extending into the extracellular vestibule, and locks the transporter in an outward open conformation. Our findings propose mechanisms for the non-competitive inhibition of DAT and attenuation of cocaine potency by AC-4-248 and provide a basis for the rational design of more efficacious atypical inhibitors. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9eup.cif.gz 9eup.cif.gz | 366.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9eup.ent.gz pdb9eup.ent.gz | 297.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9eup.json.gz 9eup.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eu/9eup https://data.pdbj.org/pub/pdb/validation_reports/eu/9eup ftp://data.pdbj.org/pub/pdb/validation_reports/eu/9eup ftp://data.pdbj.org/pub/pdb/validation_reports/eu/9eup | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  19979MC  9euoC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 60939.520 Da / Num. of mol.: 1 / Mutation: V74A, V275A, V311A, L415A, G538L Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: Q7K4Y6 Homo sapiens (human) / References: UniProt: Q7K4Y6 |
|---|
-Antibody , 2 types, 2 molecules HL
| #2: Antibody | Mass: 25921.338 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #3: Antibody | Mass: 25840.607 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Non-polymers , 5 types, 6 molecules 
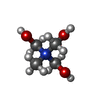







| #4: Chemical | ChemComp-Y01 / | ||
|---|---|---|---|
| #5: Chemical | ChemComp-144 / | ||
| #6: Chemical | ChemComp-CLR / | ||
| #7: Chemical | | #8: Chemical | ChemComp-CL / | |
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Inhibitor-free Drosophila melanogaster dopamine transporter in complex with Fab 9D5 Type: COMPLEX / Entity ID: #1-#3 / Source: RECOMBINANT | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.107602 MDa / Experimental value: NO | |||||||||||||||||||||||||
| Source (natural) | Organism:  | |||||||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||
| Buffer solution | pH: 8 | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Conc.: 2.3 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid type: C-flat-1.2/1.3 | |||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 59.2 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 4294 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 137600 / Symmetry type: POINT | ||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 4M48 Accession code: 4M48 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj














