[English] 日本語
 Yorodumi
Yorodumi- EMDB-19885: Low-dose cryo-electron ptychographic reconstruction of TMV record... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Low-dose cryo-electron ptychographic reconstruction of TMV recorded with CSA of 6.1 mrad | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid / Tobacco Mosaic Virus / VIRUS | |||||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | |||||||||
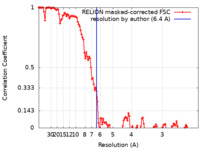
| Method | helical reconstruction / cryo EM / Resolution: 6.4 Å | |||||||||
 Authors Authors | Mohammed I / Stalhberg H / Kucukoglu B | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution. Authors: Berk Küçükoğlu / Inayathulla Mohammed / Ricardo C Guerrero-Ferreira / Stephanie M Ribet / Georgios Varnavides / Max Leo Leidl / Kelvin Lau / Sergey Nazarov / Alexander Myasnikov / ...Authors: Berk Küçükoğlu / Inayathulla Mohammed / Ricardo C Guerrero-Ferreira / Stephanie M Ribet / Georgios Varnavides / Max Leo Leidl / Kelvin Lau / Sergey Nazarov / Alexander Myasnikov / Massimo Kube / Julika Radecke / Carsten Sachse / Knut Müller-Caspary / Colin Ophus / Henning Stahlberg /    Abstract: Cryo-transmission electron microscopy (cryo-EM) of frozen hydrated specimens is an efficient method for the structural analysis of purified biological molecules. However, cryo-EM and cryo-electron ...Cryo-transmission electron microscopy (cryo-EM) of frozen hydrated specimens is an efficient method for the structural analysis of purified biological molecules. However, cryo-EM and cryo-electron tomography are limited by the low signal-to-noise ratio (SNR) of recorded images, making detection of smaller particles challenging. For dose-resilient samples often studied in the physical sciences, electron ptychography - a coherent diffractive imaging technique using 4D scanning transmission electron microscopy (4D-STEM) - has recently demonstrated excellent SNR and resolution down to tens of picometers for thin specimens imaged at room temperature. Here we apply 4D-STEM and ptychographic data analysis to frozen hydrated proteins, reaching sub-nanometer resolution 3D reconstructions. We employ low-dose cryo-EM with an aberration-corrected, convergent electron beam to collect 4D-STEM data for our reconstructions. The high frame rate of the electron detector allows us to record large datasets of electron diffraction patterns with substantial overlaps between the interaction volumes of adjacent scan positions, from which the scattering potentials of the samples are iteratively reconstructed. The reconstructed micrographs show strong SNR enabling the reconstruction of the structure of apoferritin protein at up to 5.8 Å resolution. We also show structural analysis of the Phi92 capsid and sheath, tobacco mosaic virus, and bacteriorhodopsin at slightly lower resolutions. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution Authors: Mohammed I / Stalhberg H / Kucukoglu B | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19885.map.gz emd_19885.map.gz | 15.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19885-v30.xml emd-19885-v30.xml emd-19885.xml emd-19885.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19885_fsc.xml emd_19885_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_19885.png emd_19885.png | 73 KB | ||
| Masks |  emd_19885_msk_1.map emd_19885_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19885.cif.gz emd-19885.cif.gz | 4.4 KB | ||
| Others |  emd_19885_half_map_1.map.gz emd_19885_half_map_1.map.gz emd_19885_half_map_2.map.gz emd_19885_half_map_2.map.gz | 118.7 MB 118.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19885 http://ftp.pdbj.org/pub/emdb/structures/EMD-19885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19885 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19885.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19885.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1659 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19885_msk_1.map emd_19885_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19885_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19885_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tobacco mosaic virus
| Entire | Name:   Tobacco mosaic virus Tobacco mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1: Tobacco mosaic virus
| Supramolecule | Name: Tobacco mosaic virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 12242 / Sci species name: Tobacco mosaic virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Molecular weight | Theoretical: 40.0 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DECTRIS ELA (1k x 0.5k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Cs: 0.1 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)