+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Oligomeric structure of SynDLP in presence of GTP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BDLP / cyanobacteria / membrane remodeling / GTPase / LIPID BINDING PROTEIN | |||||||||
| Function / homology | Mitofusin family / Dynamin, N-terminal / Dynamin family / GTPase activity / GTP binding / P-loop containing nucleoside triphosphate hydrolase / membrane / Slr0869 protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Junglas B / Gewehr L / Schoennenbeck P / Schneider D / Sachse C | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP. Authors: Benedikt Junglas / Lucas Gewehr / Lara Mernberger / Philipp Schönnenbeck / Ruven Jilly / Nadja Hellmann / Dirk Schneider / Carsten Sachse /  Abstract: SynDLP, a dynamin-like protein (DLP) encoded in the cyanobacterium Synechocystis sp. PCC 6803, has recently been identified to be structurally highly similar to eukaryotic dynamins. To elucidate ...SynDLP, a dynamin-like protein (DLP) encoded in the cyanobacterium Synechocystis sp. PCC 6803, has recently been identified to be structurally highly similar to eukaryotic dynamins. To elucidate structural changes during guanosine triphosphate (GTP) hydrolysis, we solved the cryoelectron microscopy (cryo-EM) structures of oligomeric full-length SynDLP after addition of guanosine diphosphate (GDP) at 4.1 Å and GTP at 3.6-Å resolution as well as a GMPPNP-bound dimer structure of a minimal G-domain construct of SynDLP at 3.8-Å resolution. In comparison with what has been seen in the previously resolved apo structure, we found that the G-domain is tilted upward relative to the stalk upon GTP hydrolysis and that the G-domain dimerizes via an additional extended dimerization domain not present in canonical G-domains. When incubated with lipid vesicles, we observed formation of irregular tubular SynDLP assemblies that interact with negatively charged lipids. Here, we provide the structural framework of a series of different functional SynDLP assembly states during GTP turnover. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19812.map.gz emd_19812.map.gz | 171.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19812-v30.xml emd-19812-v30.xml emd-19812.xml emd-19812.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19812_fsc.xml emd_19812_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_19812.png emd_19812.png | 67.9 KB | ||
| Filedesc metadata |  emd-19812.cif.gz emd-19812.cif.gz | 5.5 KB | ||
| Others |  emd_19812_half_map_1.map.gz emd_19812_half_map_1.map.gz emd_19812_half_map_2.map.gz emd_19812_half_map_2.map.gz | 322.1 MB 322.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19812 http://ftp.pdbj.org/pub/emdb/structures/EMD-19812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19812 | HTTPS FTP |
-Related structure data
| Related structure data |  9em7MC  9em8C  9em9C  15695 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19812.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19812.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.869 Å | ||||||||||||||||||||||||||||||||||||
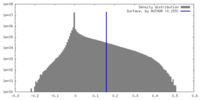
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : filamentous homo-oligomer of SynDLP in presence of GTP
| Entire | Name: filamentous homo-oligomer of SynDLP in presence of GTP |
|---|---|
| Components |
|
-Supramolecule #1: filamentous homo-oligomer of SynDLP in presence of GTP
| Supramolecule | Name: filamentous homo-oligomer of SynDLP in presence of GTP type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Slr0869 protein
| Macromolecule | Name: Slr0869 protein / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 93.705398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKIAPQCQN LREQVNQLIE LLRQEPTLRS QQDTSIVETA LGKALSPRFE IVFAGAFSAG KSMLINALLE RELLYSAEGH ATGTECHIE YANANEERVV LTFLSEAEIR QQALILAKYL NVNVGDLNIN QPEAVKVVSQ YCQKIIAEEG GENKSERAKQ A NALHLLLI ...String: MSKIAPQCQN LREQVNQLIE LLRQEPTLRS QQDTSIVETA LGKALSPRFE IVFAGAFSAG KSMLINALLE RELLYSAEGH ATGTECHIE YANANEERVV LTFLSEAEIR QQALILAKYL NVNVGDLNIN QPEAVKVVSQ YCQKIIAEEG GENKSERAKQ A NALHLLLI GFEQNRERIN TVQNSTYSMD QLNFSSLAEA AGYARRGANS AVLKRLDYFC NHSLLKDGNV LVDLPGIDAP VK EDAERAY RKIESPDTSA VICVLKPAAA GDMSAEETQL LERISKNHGI RDRVFYVFNR IDDTWYNTQL RQRLEGLIQS QFR DNSRVY KTSGLLGFYG SQVKQTNSST RFGLDSIFAT TIKGFDGEEE TPQFVSEFNN YCANSGKLLS TAFRVSVNGY ETSN ENYVR ILSEWGIPLV DQLIHDSGIE SFRSGIGLYL AEEKYPELFA TLANDLQPLC IALRQFYLEN YRQLDSQPRE IAAMK AQEL TLLNQEMQNL GIEFKKYMSA QINDVVIGND REFDQDFTKL KARMVARLDE LLKTFSVMNA YKRATESHPR NSTAPF IAV LVEALYYLAN ELEDAFIEAI HELVKNFFQR LGDRLRKVDC YHQVYRLVGN DGGIEQLLRR AEEDITKALV NEARTEC DR YVRESPRFYD EGTFSIYQFR QTLQQTSQGY DAQAIVEAEP AIKELLKLDF EPKVFNTVRK NFRQTVNNTL KTHLLPMA E EQAQIILEQY DVARKYREQT LEQDAEEKIA RNSRLQSEIK QKIDLYQTSI VSINECLKAM QIFEQLPVIT ESDITKQAE IVADADFVEI VELEHHHHHH UniProtKB: Slr0869 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 44.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)