[English] 日本語
 Yorodumi
Yorodumi- EMDB-19738: Full-length human cystathionine beta-synthase, basal state, singl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full-length human cystathionine beta-synthase, basal state, single particle reconstruction | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Filament / Allostery / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationCysteine formation from homocysteine / homocysteine catabolic process / cystathionine beta-synthase / modified amino acid binding / cystathionine beta-synthase activity / L-serine catabolic process / regulation of nitric oxide mediated signal transduction / Metabolism of ingested SeMet, Sec, MeSec into H2Se / cysteine biosynthetic process via cystathionine / carbon monoxide binding ...Cysteine formation from homocysteine / homocysteine catabolic process / cystathionine beta-synthase / modified amino acid binding / cystathionine beta-synthase activity / L-serine catabolic process / regulation of nitric oxide mediated signal transduction / Metabolism of ingested SeMet, Sec, MeSec into H2Se / cysteine biosynthetic process via cystathionine / carbon monoxide binding / hydrogen sulfide biosynthetic process / L-serine metabolic process / homocysteine metabolic process / cartilage development involved in endochondral bone morphogenesis / L-cysteine catabolic process / cerebellum morphogenesis / cysteine biosynthetic process / response to folic acid / endochondral ossification / transsulfuration / cysteine biosynthetic process from serine / nitric oxide binding / DNA protection / S-adenosyl-L-methionine binding / nitrite reductase (NO-forming) activity / regulation of JNK cascade / superoxide metabolic process / blood vessel remodeling / maternal process involved in female pregnancy / blood vessel diameter maintenance / oxygen binding / pyridoxal phosphate binding / cellular response to hypoxia / heme binding / ubiquitin protein ligase binding / negative regulation of apoptotic process / enzyme binding / protein homodimerization activity / metal ion binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
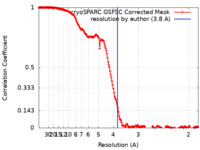
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | McCorvie TJ / Yue WW | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Architecture and regulation of filamentous human cystathionine beta-synthase. Authors: Thomas J McCorvie / Douglas Adamoski / Raquel A C Machado / Jiazhi Tang / Henry J Bailey / Douglas S M Ferreira / Claire Strain-Damerell / Arnaud Baslé / Andre L B Ambrosio / Sandra M G Dias / Wyatt W Yue /    Abstract: Cystathionine beta-synthase (CBS) is an essential metabolic enzyme across all domains of life for the production of glutathione, cysteine, and hydrogen sulfide. Appended to the conserved catalytic ...Cystathionine beta-synthase (CBS) is an essential metabolic enzyme across all domains of life for the production of glutathione, cysteine, and hydrogen sulfide. Appended to the conserved catalytic domain of human CBS is a regulatory domain that modulates activity by S-adenosyl-L-methionine (SAM) and promotes oligomerisation. Here we show using cryo-electron microscopy that full-length human CBS in the basal and SAM-bound activated states polymerises as filaments mediated by a conserved regulatory domain loop. In the basal state, CBS regulatory domains sterically block the catalytic domain active site, resulting in a low-activity filament with three CBS dimers per turn. This steric block is removed when in the activated state, one SAM molecule binds to the regulatory domain, forming a high-activity filament with two CBS dimers per turn. These large conformational changes result in a central filament of SAM-stabilised regulatory domains at the core, decorated with highly flexible catalytic domains. Polymerisation stabilises CBS and reduces thermal denaturation. In PC-3 cells, we observed nutrient-responsive CBS filamentation that disassembles when methionine is depleted and reversed in the presence of SAM. Together our findings extend our understanding of CBS enzyme regulation, and open new avenues for investigating the pathogenic mechanism and therapeutic opportunities for CBS-associated disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19738.map.gz emd_19738.map.gz | 450.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19738-v30.xml emd-19738-v30.xml emd-19738.xml emd-19738.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19738_fsc.xml emd_19738_fsc.xml | 16.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_19738.png emd_19738.png | 48.8 KB | ||
| Masks |  emd_19738_msk_1.map emd_19738_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19738.cif.gz emd-19738.cif.gz | 6.5 KB | ||
| Others |  emd_19738_additional_1.map.gz emd_19738_additional_1.map.gz emd_19738_half_map_1.map.gz emd_19738_half_map_1.map.gz emd_19738_half_map_2.map.gz emd_19738_half_map_2.map.gz | 237.9 MB 442.8 MB 442.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19738 http://ftp.pdbj.org/pub/emdb/structures/EMD-19738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19738 | HTTPS FTP |
-Validation report
| Summary document |  emd_19738_validation.pdf.gz emd_19738_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19738_full_validation.pdf.gz emd_19738_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_19738_validation.xml.gz emd_19738_validation.xml.gz | 25.5 KB | Display | |
| Data in CIF |  emd_19738_validation.cif.gz emd_19738_validation.cif.gz | 33.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19738 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19738 | HTTPS FTP |
-Related structure data
| Related structure data |  8s5kMC  8s5hC  8s5iC  8s5jC  8s5lC  8s5mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19738.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19738.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.934 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19738_msk_1.map emd_19738_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened map
| File | emd_19738_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19738_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19738_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helical assembly of full-length human cystathionine beta-synthase...
| Entire | Name: Helical assembly of full-length human cystathionine beta-synthase in the basal state |
|---|---|
| Components |
|
-Supramolecule #1: Helical assembly of full-length human cystathionine beta-synthase...
| Supramolecule | Name: Helical assembly of full-length human cystathionine beta-synthase in the basal state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cystathionine beta-synthase
| Macromolecule | Name: Cystathionine beta-synthase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: cystathionine beta-synthase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.980578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMPSETPQAE VGPTGCPHRS GPHSAKGSLE KGSPEDKEAK EPLWIRPDAP SRCTWQLGRP ASESPHHHTA PAKSPKILPD ILKKIGDTP MVRINKIGKK FGLKCELLAK CEFFNAGGSV (LLP)DRISLRMIE DAERDGTLKP GDTIIEPTSG NTGIGLA LA AAVRGYRCII ...String: SMPSETPQAE VGPTGCPHRS GPHSAKGSLE KGSPEDKEAK EPLWIRPDAP SRCTWQLGRP ASESPHHHTA PAKSPKILPD ILKKIGDTP MVRINKIGKK FGLKCELLAK CEFFNAGGSV (LLP)DRISLRMIE DAERDGTLKP GDTIIEPTSG NTGIGLA LA AAVRGYRCII VMPEKMSSEK VDVLRALGAE IVRTPTNARF DSPESHVGVA WRLKNEIPNS HILDQYRNAS NPLAHYDT T ADEILQQCDG KLDMLVASVG TGGTITGIAR KLKEKCPGCR IIGVDPEGSI LAEPEELNQT EQTTYEVEGI GYDFIPTVL DRTVVDKWFK SNDEEAFTFA RMLIAQEGLL CGGSAGSTVA VAVKAAQELQ EGQRCVVILP DSVRNYMTKF LSDRWMLQKG FLKEEDLTE KKPWWWHLRV QELGLSAPLT VLPTITCGHT IEILREKGFD QAPVVDEAGV ILGMVTLGNM LSSLLAGKVQ P SDQVGKVI YKQFKQIRLT DTLGRLSHIL EMDHFALVVH EQIQYHSTGK SSQRQMVFGV VTAIDLLNFV AAQERDQK UniProtKB: Cystathionine beta-synthase |
-Macromolecule #2: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 2 / Number of copies: 8 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: filter sterile 25 mM HEPES, pH 7.5, 200 mM NaCl, 2.0 mM TCEP, 0.005% (v/v) tween-20 | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2628 / Average exposure time: 5.18 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)